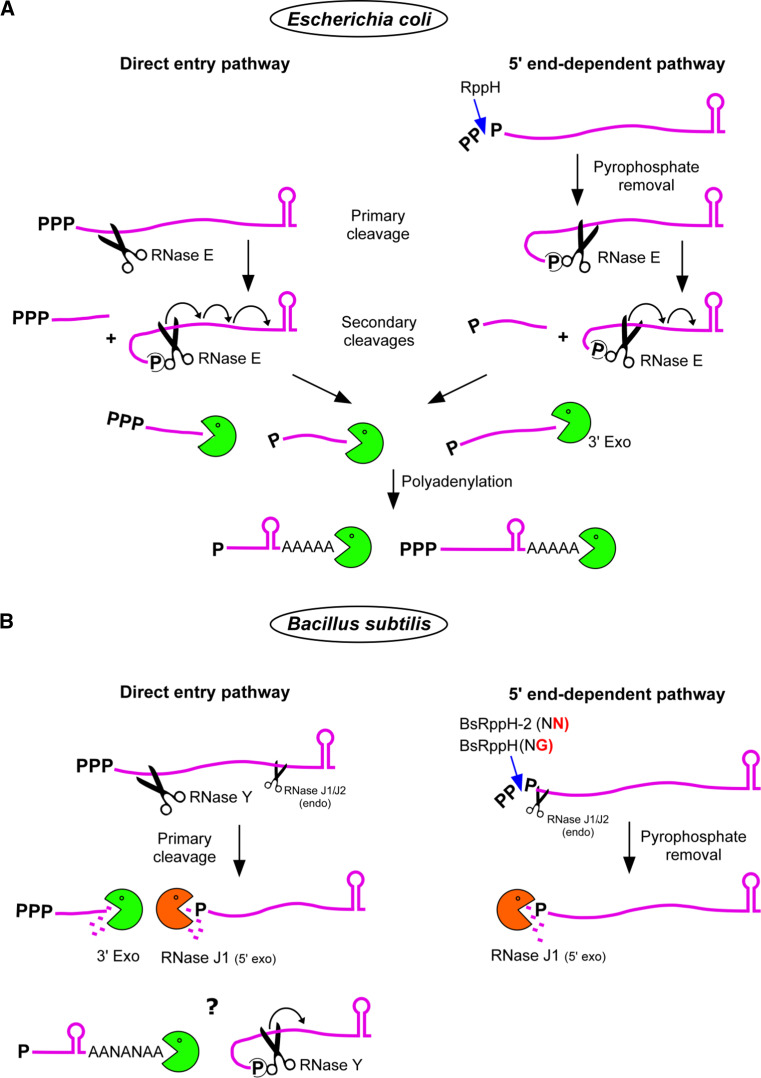
Fig. 3.
RNA degradation pathways in E. coli and B. subtilis. Initiation of mRNA decay as defined by the first nucleolytic cleavage can depend on a variety of parameters that render a given mRNA susceptible to the action of an RNase (e.g., translation efficiency, 5′ end conversion, stochastic events, etc., see text). a In E. coli, the major direct entry pathway involves a primary cleavage of the native transcript by RNase E. The upstream fragments are rapidly degraded by 3′ exoribonucleases (RNase II, PNPase, RNase R, and oligoribonuclease for short oligonucleotides). The 5′ monophosphorylated downstream fragment is preferentially recognized by the 5′ sensor of RNase E, which enhances the rate of subsequent cleavages (>20-fold, at least in vitro). This causes a wave of secondary downstream cleavages proceeding in a 5′–3′ direction each generating a 3′-OH upstream fragment that is degraded by 3′ exonucleases. Decay intermediates whether or not protected by 3′ secondary structure can be polyadenylated by poly(A) polymerase, enabling the 3′ exonucleases to re-engage several times if necessary to produce complete degradation (see main text). Polyadenylation can also be observed on full-length transcripts containing the transcription terminator (not shown in the figure) but does not represent a major pathway to initiate mRNA decay (see main text, “The 3′ end: tailing, scavenging and surveillance”). A second pathway of mRNA degradation in E. coli is 5′ end-dependent and starts with pyrophosphate removal by the pyrophosphohydrolase RppH. This tethers RNase E to the 5′ end of the transcript and stimulates downstream cleavage in the same way as described for secondary cleavages above. Refer to the legend of Fig. 1 and text. b In B. subtilis, the pathways initiating mRNA decay are similar to E. coli but the players are different. In the major direct entry pathway, the primary cleavage is affected by RNase Y and to a lesser extent by RNase J1/J2 or another endonuclease. The upstream fragments are degraded mainly by PNPase, in contrast to RNase II in E. coli [313–315]. The monophosphorylated downstream cleavage products are degraded 5′–3′ by RNase J1/J2 in exonuclease mode and can proceed to the 3′ end. It is interesting to note that the B. subtilis extracts used to demonstrate the largely phosphorolytic degradation of RNA to mononucleotides [313] most likely did not measure the contribution of the, at the time, unknown hydrolytic RNases J1/J2 to exonucleolytic decay, due to the 5′ triphosphorylated RNA substrate used and the fact that most of the ribosome associated RNase J was probably eliminated during extract preparation [313]. B. subtilis has no poly(A) polymerase but A-rich polynucleotide tails synthesized by an unknown enzyme (indicated by an ANA sequence) are found essentially on degradation intermediates [138]. The question mark indicates that it is not clear whether they contribute to the degradation of 3′ structured fragments. However, 3′ terminal fragments containing the transcription terminator are very resistant to 3′ exonuclease attack. The 5′ exonuclease activity of RNase J is thus very useful to degrade 3′ structured RNA fragments. Similar to E. coli, conversion of the native 5′ PPP to a 5′ P by BsRppH (which prefers a G in second position, see text) and BsRppH-2 (not yet identified, but insensitive to N-terminal sequence, see text) renders the mRNA susceptible to the 5′ exonuclease activity of RNase J. In vitro, RNase J can also cleave endonucleolytically a native transcript close to the 5′ end probably by threading the 5′ PPP through the RNA entry channel and past the 5′ P binding pocket. It is not known whether this “sliding endonuclease” mode plays a significant role in 5′ end conversion in vivo. Similarly, RNase Y activity is stimulated by a 5′ P group in much the same way as RNase E, but to what extent RNase Y competes with RNase J for binding to a monophosphorylated 5′ end in vivo remains to be analyzed (indicated by a question mark)