Figure 6.
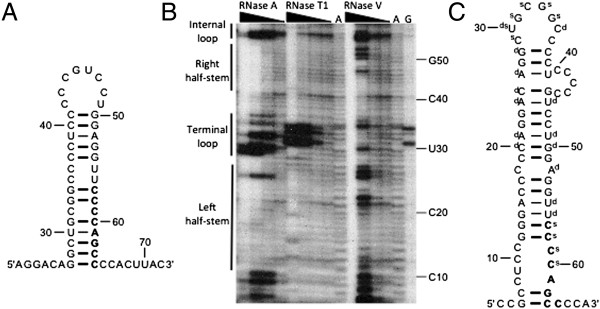
High-order structure of the ζ-globin 3′UTR. (A) A thermodynamically favored structure resulting from unrefined m-fold analysis of the full-length ζ-globin 3′UTR (detail). The ZMRE is indicated in boldface. Double and triple hydrogen bond interactions are indicated by thin and thick connectors, respectively. (B) Enzymatic secondary-structure mapping of the ζ-globin 3′UTR (detail). 5′-end [32P]-labeled RNAs corresponding to the ζ-globin 3′UTR (and contiguous 18-nt polyadenylate tail) were digested with RNases A, T1, and V1 at four different concentrations (wedges), then resolved on an acrylamide-urea gel. Nucleotide assignments (right) were deduced by aligning the known 3′UTR sequence to specific bands corresponding to guanine bases (lane G), which were generated by RNase T1 digestion of denatured 3′UTRs. An alkaline hydrolysis ladder (lanes A) provides additional sequence information at single-nucleotide resolution. Regions of 3′UTR that exhibit stem- and loop-like characteristics (i.e., sensitivities to double- and single-strand-specific nucleases, respectively) are indicated to the left. (C) Thermodynamically favored structure resulting from m-fold analysis of the full-length ζ-globin 3′UTR, where base-pairing is enforced for nts 20–21, 24–26, and 38; and prohibited for nts 29–34 (detail). Nucleotides that displayed less-exacting single- or double-strand characteristics were not used for predictive purposes but are indicated by s and d, respectively.
