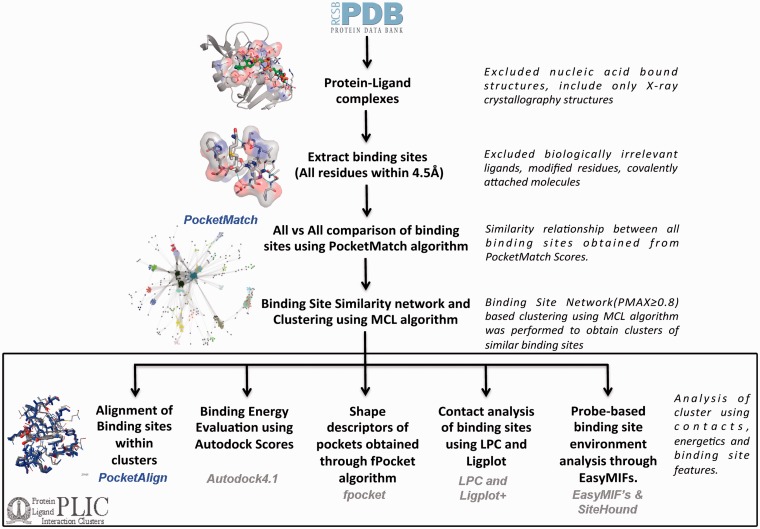
Figure 1.
PLIC database workflow. The flowchart illustrates the different steps involved in the construction of the PLIC database. All the protein–ligand complexes are downloaded from the PDB, and binding sites (comprising all the residues that are within 4.5 Å of any ligand atom) are extracted. Only the biologically relevant ligands are selected that resulted in 84 846 binding sites. An exhaustive all-versus-all comparison of these 84 846 binding sites is performed using PocketMatch, and a binding site similarity network is constructed at a PMAX cutoff of 0.8. Network-based clustering of binding sites is performed using the MCL algorithm to obtain clusters of similar binding sites. All the different attributes that are calculated for the interactions within the clusters along with computational tools that were used to derive them are mentioned in the box.