Fig. 2.
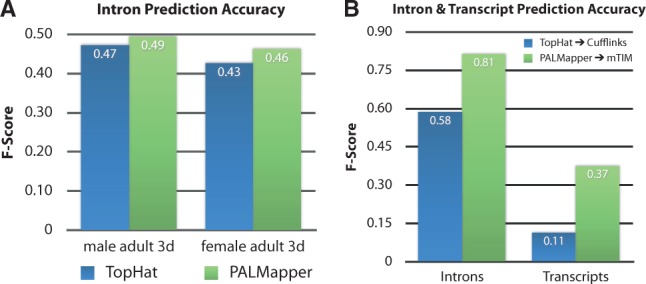
(A) Performance comparison of two alignment programs integrated in Oqtans, evaluated on the data from the use case in terms of intron accuracy (see Supplementary Fig. S3 for details). Such comparative evaluations are made easy, since the replicability assertion of the Galaxy Oqtans setup ensures otherwise identical comparisons. (B) Performance comparison from Görnitz et al. (2011), where PALMapper and TopHat alignments are processed with the de novo transcript inference tools mTIM and Cufflinks, again demonstrating the value of Oqtans for comparisons of analysis tool
