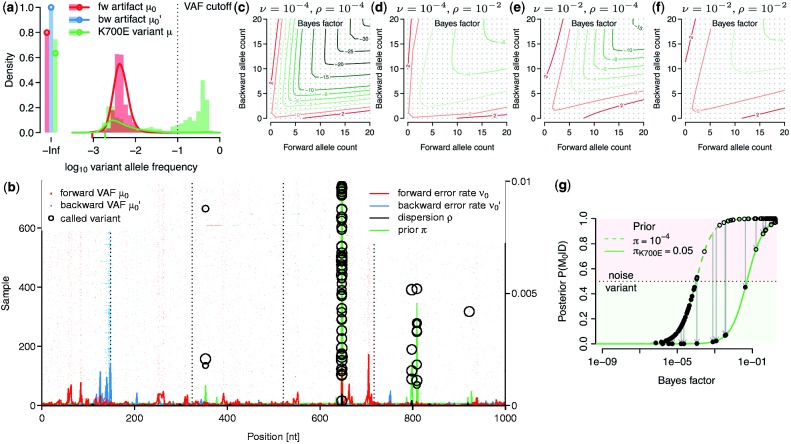
Fig. 1.
General illustration of our approach. (a) Distribution of observed and expected VAFs across samples. The histograms denote the VAF 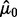 and
and 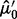 of a recurrent artifact occurring at low frequencies in ∼20% of the samples in forward, but not in the reverse orientation. The solid lines denote the expected distribution based on a beta-binomial model, Equation (1), with mean
of a recurrent artifact occurring at low frequencies in ∼20% of the samples in forward, but not in the reverse orientation. The solid lines denote the expected distribution based on a beta-binomial model, Equation (1), with mean 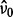 and
and 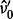 defined as the average across all samples with VAF
defined as the average across all samples with VAF 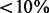 . The third histogram denotes the SF3B1 K700E variant present at clonal and subclonal frequencies, with the curve denoting the expected frequency distribution. (b) Heatmap of 1000 nt from five adjacent bait sets targeting the SF3B1 gene in 683 samples. The intensity of each pixel represents VAF of cytosine,
. The third histogram denotes the SF3B1 K700E variant present at clonal and subclonal frequencies, with the curve denoting the expected frequency distribution. (b) Heatmap of 1000 nt from five adjacent bait sets targeting the SF3B1 gene in 683 samples. The intensity of each pixel represents VAF of cytosine, 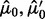 , in a given sample (y, left axis) and position (x). If the relative frequency is identical, pixels tend to be black. Curves on the bottom indicate the error rates
, in a given sample (y, left axis) and position (x). If the relative frequency is identical, pixels tend to be black. Curves on the bottom indicate the error rates 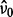 and
and 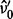 in forward and reverse directions (right y-axis). The black line is the estimated dispersion
in forward and reverse directions (right y-axis). The black line is the estimated dispersion 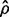 . The prior π of finding a true variant is derived from the COSMIC database. Circles are drawn around variants with a posterior
. The prior π of finding a true variant is derived from the COSMIC database. Circles are drawn around variants with a posterior 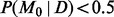 ; the area of each circle is proportional to the VAF. At position 650 resides the K700E hotspot mutation with many variant calls. (c–f) Bayes factors [Equation (7)] as a function of forward (x) and reverse (y) allele counts for different error rates
; the area of each circle is proportional to the VAF. At position 650 resides the K700E hotspot mutation with many variant calls. (c–f) Bayes factors [Equation (7)] as a function of forward (x) and reverse (y) allele counts for different error rates 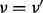 and dispersions
and dispersions 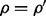 . (g) A variant-specific prior π influences the Bayes factor needed to call a variant at a given cutoff on the posterior probability, Equation (9)
. (g) A variant-specific prior π influences the Bayes factor needed to call a variant at a given cutoff on the posterior probability, Equation (9)