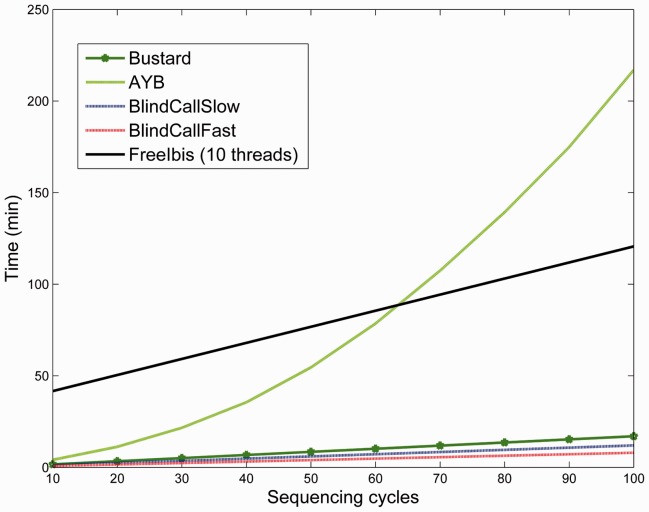
Fig. 5.
Base-calling by blind deconvolution is scalable to long read lengths. We compare the computational time of BlindCall with a state-of-the-art probabilistic base caller AYB, the state-of-the-art supervised learning method freeIbis and Illumina’s Bustard on the PhiX174 dataset reported in Table 1 as a function of the number of sequencing cycles. Since most model-based base callers resort to a dynamic programming solution, running time is quadratic with respect to the read length. In contrast, BlindCall scales linearly with read length. Base callers based on the blind deconvolution framework will be able to scale as sequencers produce longer reads. freeIbis also scales linearly but is much slower than BlindCall