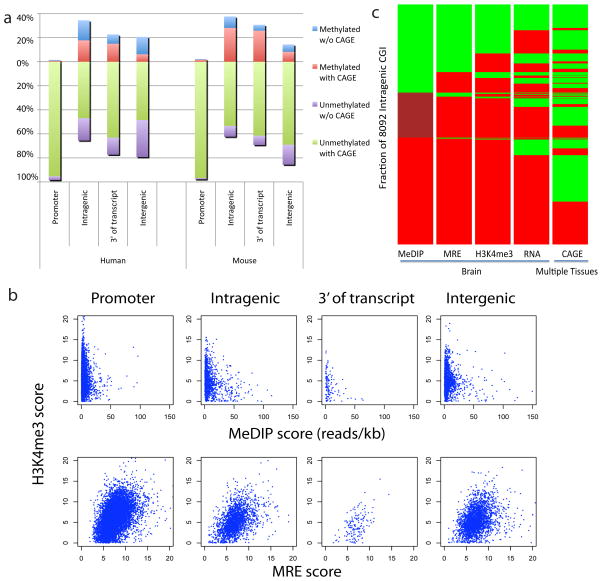
Figure 2. Differentially methylated intragenic CGIs exhibit features of promoters.
a, Methylated CGIs are indicated above the zero line and unmethylated CGIs are below. For human, the methylation data is from frontal cortex, and CAGE tags are derived from multiple tissues11,23. For mouse, the methylation data includes the same set of tissues described in figure 1c, and CAGE data are derived from multiple mouse tissues11,12. 91% of human intragenic CGI CAGE tags mapped outside of exons and are probably not derived from posttranscriptional processing. b, H3K4me3 tissue-ChIP-seq normalized internal coverage (NIC) scores compared to MeDIP- and MRE-seq methylation data at CGIs for human frontal cortex. c, Heatmap view of the status of 8092 intragenic CGIs based on five genome-wide datasets. Each island is coloured according to its status and sorted from top to bottom in the order of increasing signal in MeDIP-seq, then within the three MeDIP-defined subgroups by signals in MRE-seq. This process is performed iteratively based on H3K4me3, RNA-seq TSS and CAGE status. For MeDIP-seq, green indicates unmethylated (0–20 reads/kb), maroon indicates partially methylated (20–50 reads/kb), and red indicates methylated (>50 reads/kb); For MRE-seq, green indicates unmethylated (MRE score 0–5), red indicates methylated (MRE score >5); For H3K4me3 ChIP-seq, green indicates active/with signal, red indicates inactive/without signal. For RNA-seq TSS, green indicates evidence for TSS, red indicates lack of evidence for TSS (see Supplemental Methods). For CAGE, green indicates CAGE tags from one or more tissues that overlap the CGI; red indicates lack of overlapping CAGE tags.