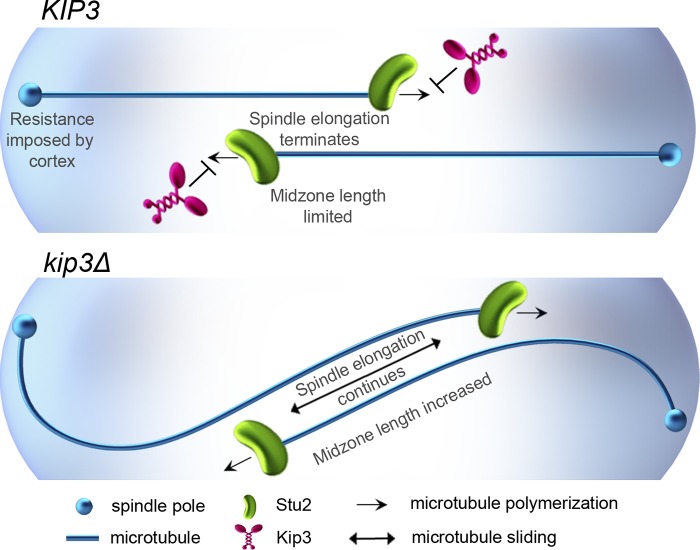
The depolymerase activity of Kip3 suppresses spindle microtubule polymerization to limit spindle midzone length and prevent microtubule buckling in response to sliding forces.
Abstract
Mitotic spindle function is critical for cell division and genomic stability. During anaphase, the elongating spindle physically segregates the sister chromatids. However, the molecular mechanisms that determine the extent of anaphase spindle elongation remain largely unclear. In a screen of yeast mutants with altered spindle length, we identified the kinesin-8 Kip3 as essential to scale spindle length with cell size. Kip3 is a multifunctional motor protein with microtubule depolymerase, plus-end motility, and antiparallel sliding activities. Here we demonstrate that the depolymerase activity is indispensable to control spindle length, whereas the motility and sliding activities are not sufficient. Furthermore, the microtubule-destabilizing activity is required to counteract Stu2/XMAP215-mediated microtubule polymerization so that spindle elongation terminates once spindles reach the appropriate final length. Our data support a model where Kip3 directly suppresses spindle microtubule polymerization, limiting midzone length. As a result, sliding forces within the midzone cannot buckle spindle microtubules, which allows the cell boundary to define the extent of spindle elongation.
Introduction
During cell division, the mitotic spindle undergoes continuous dynamic changes yet maintains the structural integrity required to ensure genomic stability (Goshima and Scholey, 2010). During anaphase, spindles must obtain a length sufficient to partition the chromosomes. In addition, a stable bipolar structure is required to restrain separated chromatids in each daughter cell before cytokinesis, allow for spindle positioning during asymmetric divisions, and facilitate cytokinesis across the spindle midbody (Roostalu et al., 2010; Fededa and Gerlich, 2012; McNally, 2013). Anaphase spindle length has been observed to scale with cell size to various degrees in diverse organisms or cell types (Storchová et al., 2006; Hara and Kimura, 2009; Hu et al., 2011; Kotadia et al., 2012). Therefore, mechanisms likely exist that regulate the dynamics, elongation, and length of spindles during anaphase. However, how anaphase spindles scale with cell size and how elongation is terminated when spindles reach proper length remain open questions.
Structural stability of the bipolar anaphase spindle depends largely on the midzone, where antiparallel microtubules emanating from each spindle pole overlap. Midzone microtubules maintain association through cross-linking proteins of the Ase1–PRC1–MAP65 family, while kinesin-5 motors, such as Cin8 and Kip1, provide antiparallel sliding forces to facilitate spindle elongation (Straight et al., 1998; Schuyler et al., 2003). To preserve structural integrity, the dynamics of these overlapping microtubules must be controlled so that overall polymerization accompanies spindle elongation while net depolymerization of the antiparallel overlap region remains limited.
Previous work in the budding yeast Saccharomyces cerevisiae identified a group of mutants with slightly increased anaphase spindle length, and it was postulated that these mutants were defective in spindle disassembly (Straight et al., 1998; Buvelot et al., 2003; Vizeacoumar et al., 2010). We screened these mutants for failure to terminate spindle elongation during anaphase and identified the conserved kinesin-8, Kip3, as crucial for regulating anaphase spindle elongation and properly matching spindle length with cell length. Kinesin-8s are a conserved family of plus end–directed microtubule motors that localize to microtubule plus-ends in vivo and regulate the dynamic behavior of microtubules in vitro and in vivo (West et al., 2001; Garcia et al., 2002; Goshima and Vale, 2005; Gupta et al., 2006; Varga et al., 2006; Mayr et al., 2007; Unsworth et al., 2008; Tischer et al., 2009; Du et al., 2010; Gardner et al., 2011; Erent et al., 2012; Su et al., 2013). They localize to the midzone in human (Stumpff et al., 2008; Zhang et al., 2010), Drosophila (Goshima and Vale, 2005), fission yeast (Garcia et al., 2002; West et al., 2002), and budding yeast (Gupta et al., 2006). Increased anaphase spindle length has been observed in budding yeast and Drosophila cells lacking kinesin-8 (Cottingham and Hoyt, 1997; Straight et al., 1998; Gandhi et al., 2004; Goshima and Vale, 2005; Wang et al., 2010). Consistent with the idea that increased anaphase spindle length is a symptom of delayed disassembly, the budding yeast kinesin-8 is required for efficient spindle disassembly upon mitotic exit (Woodruff et al., 2010). However, because anaphase is typically followed closely by spindle disassembly, the role of kinesin-8 during anaphase, before disassembly, has remained obscure. Kinesin-8 is required for progression through metaphase in Drosophila S2 (Goshima and Vale, 2003) and human HeLa cells (Mayr et al., 2007), which limits study during anaphase in these systems. Conversely, budding yeast lacking kinesin-8 progress through mitosis and therefore provide an ideal system to determine the function of kinesin-8 during anaphase.
Here we show that the yeast kinesin-8, Kip3, is essential to scale the anaphase spindle with cell length. In cells lacking Kip3, spindles buckle and continue to hyper-elongate after reaching a length equal to that of the cell. Using a separation of function allele, we demonstrate that the microtubule depolymerase activity of Kip3 is required to balance Stu2-mediated polymerization and prevent spindle hyper-elongation. During anaphase, Kip3 restricts spindle microtubule polymerization and limits the region of antiparallel microtubule overlap in the midzone. Overall, our data support a model in which Kip3 limits midzone-associated elongation forces by restricting midzone length. This in turn prevents spindle elongation forces from buckling the spindle upon encountering resistance from the cell cortex, effectively terminating spindle elongation. In support of this model, inhibition of microtubule polymerization with the small molecule nocodazole restores midzone length and effectively terminates anaphase elongation in cells lacking Kip3.
Results
Kip3 is a key regulator of anaphase spindle length
In budding yeast, anaphase spindle elongation proceeds until the full-length spindle spans the length of the cell. Interestingly, when cells are challenged with a late anaphase arrest, the full-length spindle typically maintains a straight morphology and does not exceed the length of the cell (Culotti and Hartwell, 1971; Luca and Winey, 1998). To determine whether anaphase spindle length can scale with varying cell lengths, in otherwise isogenic cells, we induced a cdc15-2–mediated late anaphase arrest and determined the spindle-to-cell length ratio in GFP-tubulin–expressing cells. Within arrested cultures, cells exhibit naturally occurring variations in cell size. Our measurements revealed that spindle length was appropriately scaled with variations in cell length (Fig. 1 a), suggesting that mechanisms exist to control the extent of anaphase spindle elongation.
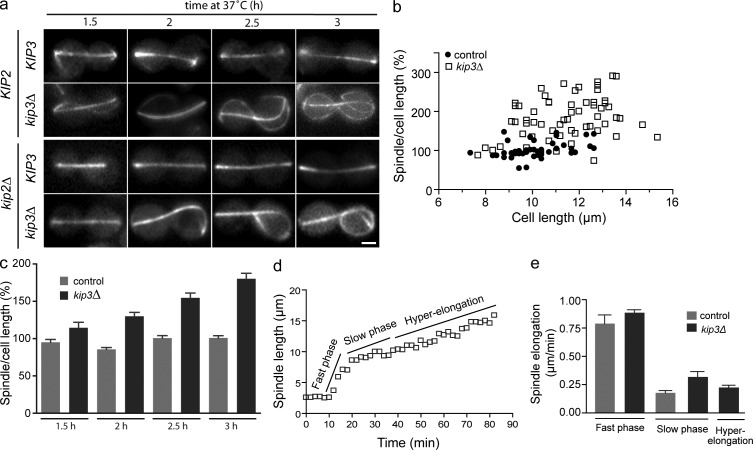
Figure 1.
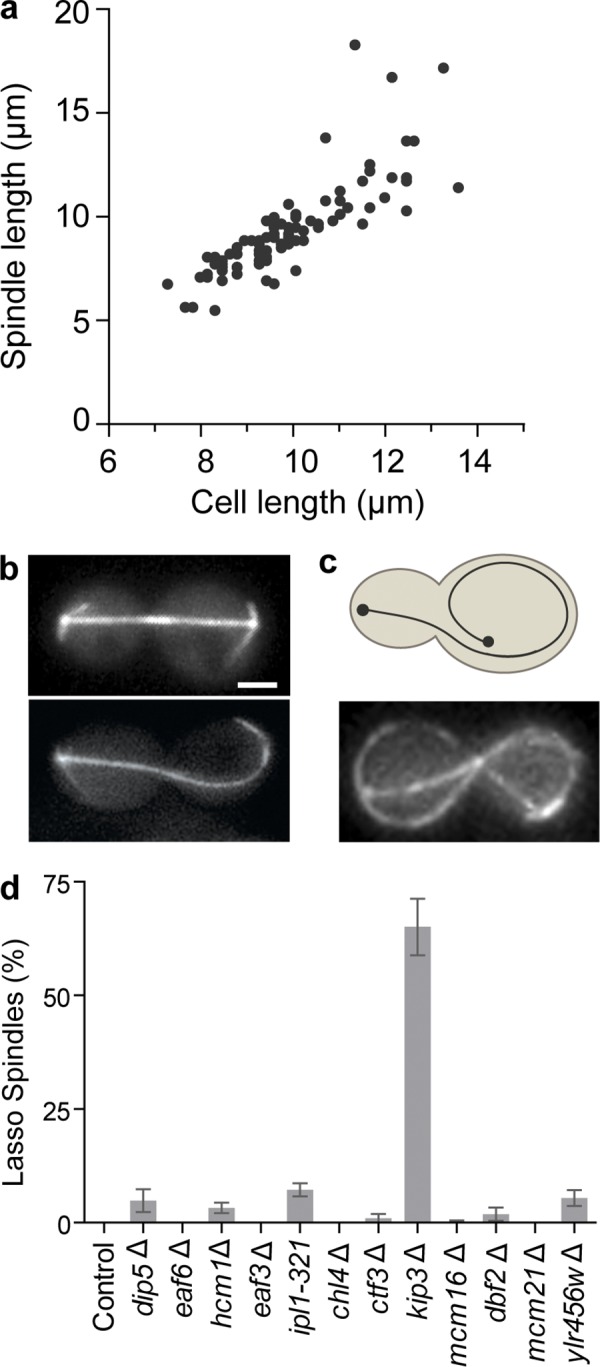
Kip3 is required to scale anaphase spindle length with cell length. (a) Spindle length was scaled with cell size in late anaphase (cdc15-2, 2 h at 37°C). The graph represents the data from two independent cultures, n = 87. (b) Representative straight (top) and fishhook (bottom) spindle images from cells expressing GFP-tubulin. (c) Schematic (top) and representative image (bottom) of the lasso spindle morphology. (d) Percentage of total anaphase spindles with lasso morphology in control and mutant cells during anaphase arrest (cdc15-2, 2.5 h at 37°C). The graph shows the average ± SE of three separate experiments. P < 0.005 for kip3Δ versus each cell type. For each cell type the compiled n = 112–540, and in each experiment n = 24–271. Bar, 2 µm.
A recent genome-wide screen of nonessential genes, plus conditional alleles of ∼100 essential genes, generated a subset of mutants that displayed an elevated level of spindles that slightly exceeded cell length (Vizeacoumar et al., 2010), a morphology termed “fishhook” (Fig. 1 b). Previous work also reported fishhook spindles in cells with mutations in the Aurora B homologue, IPL1, or the kinesin-8 KIP3 (Straight et al., 1998; Buvelot et al., 2003). The fishhook phenotype has been postulated to be a symptom of impaired spindle disassembly (Vizeacoumar et al., 2010), and indeed the products of several of these genes function in spindle disassembly (Kip3, Dbf2, Mcm21, and Ipl1; Woodruff et al., 2010). However, to determine whether any of the proteins tested regulate spindle length during anaphase, we challenged these mutant cells with a cdc15-2 arrest to prevent spindle disassembly and monitored spindle length. To identify mutants with severe loss of spindle length regulation, we scored cells for what we termed a “lasso” morphology, in which excessive spindle elongation caused the spindle to wrap along the cell periphery and double back on itself (Fig. 1 c). In anaphase-arrested control cultures we observed no lasso spindles. Strikingly, in kip3Δ cells there were 65% lasso spindles. This was at least a 10-fold increase in lasso phenotype compared with all other mutants tested (Fig. 1 d). These results clearly demonstrate that Kip3 plays a major role in regulating spindle length during anaphase, and that this function is separable from the role of Kip3 in spindle disassembly.
Kip3 terminates anaphase B elongation when spindle length matches cell size
To examine the role of Kip3 in controlling spindle length, we imaged spindles at various time points throughout the anaphase arrest. In contrast to control cdc15-2–arrested cells, spindle length in arrested kip3Δ cells clearly exceeded the cell length (Fig. 2 a). Spindles in kip3Δ cells proceeded from straight to fishhook, and then to the lasso morphology. Consistent with previous work, in kip3Δ cells astral microtubule length was significantly increased (Miller et al., 1998; Cottingham and Hoyt, 1997). The long astral microtubules often intertwined with spindle polymer and thus hindered spindle length measurements. Therefore, to reduce astral microtubule polymer we removed the kinesin Kip2 that functions to stabilize astral microtubules (Carvalho et al., 2004). Kip2 is a cytoplasmic motor that is not known to function in the spindle and, consistent with this, kip2Δ did not alter spindle elongation and morphology while allowing for direct measurement of spindle length (Fig. S1, a–c).
Figure 2.
Kip3 terminates anaphase B elongation when spindle length matches cell length. (a) Images in each row are representative of KIP2 cells ± Kip3 (top) or kip2Δ cells ± Kip3 (bottom). Cells were arrested in anaphase (cdc15-2, 37°C) for the times indicated. (b) In kip3Δ cells, proper scaling of anaphase spindle length to cell size is lost (cdc15-2, 3 h at 37°C). The graph represents the data from two independent cultures, n > 45 for each cell type. (c) Spindle length as a percentage of cell length during anaphase arrest for the indicated times. Average ± SE, n = 47–109 for each time point. P < 0.05 for kip3Δ versus control at all time points. (d) Spindle length over time in an anaphase-arrested kip3Δ cell. The graph shows a single cell from the data collected in panel e. (e) Average spindle elongation rates during anaphase in the presence and absence of Kip3. Average ± SE, n = 9–17 for each phase over at least three separate days of live imaging. P < 0.05 for slow phase. Bar, 2 µm.
Consistent with our initial observations, spindle length measurements showed that, in control cells, anaphase spindles remained scaled with cell length throughout prolonged arrest (Fig. 2 b). In contrast, the appropriate scaling of anaphase spindle length was lost in cells lacking Kip3 (Fig. 2 b). Moreover, the average spindle length observed in kip3Δ cells increased over time, revealing that spindle elongation persisted throughout the arrest (Fig. 2 c).
We next determined spindle elongation rates during anaphase arrest from time-lapse images of live cells expressing GFP-tubulin. Overall, the rate of hyper-elongation in the absence of Kip3 was comparable to normal anaphase rates (Fig. 2, d and e). In budding yeast, anaphase B consists of an initial fast phase and a second slow phase (Straight et al., 1998). After shift to restrictive temperature, the fast phase proceeded at 0.88 ± 0.03 and 0.79 ± 0.08 µm/min in kip3Δ and control cells, respectively. During the slow phase, spindles grew at 0.31 ± 0.08 relative to 0.18 ± 0.02 µm/min in kip3Δ and control cells, respectively. Similarly, the rate of spindle hyper-elongation in kip3Δ cells averaged 0.23 ± 0.02 µm/min. Together, these data reveal a novel role for Kip3 in terminating anaphase elongation when the spindle reaches a length equivalent to that of the cell.
Kip3 mediates a transition from dynamic to nondynamic spindle midzone when anaphase spindles reach full length
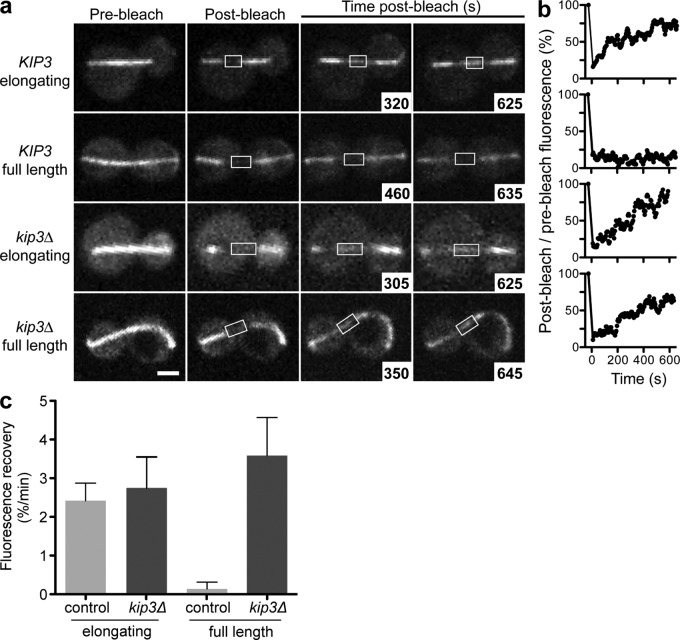
Our results reveal that in the absence of Kip3 the resistance imposed by the cell wall against the spindle poles does not limit spindle length. Therefore, a Kip3-dependent mechanism that ensures spindle elongation terminates at the appropriate length is intrinsic to the spindle. Spindle elongation in yeast is largely achieved through net polymerization and antiparallel sliding of midzone microtubules (Maddox et al., 2000). To understand how Kip3 influences midzone behavior of elongating and full-length anaphase spindles, we used FRAP to measure the overall dynamic behavior of midzone microtubules. Typically, full-length anaphase spindles become rapidly disassembled as cells progress into cytokinesis. This timing of disassembly obscures the dynamic behavior of the midzone when anaphase elongation is terminated. Therefore, to clearly monitor midzone microtubule dynamics in full-length spindles before disassembly, we used anaphase-arrested cells in the presence or absence of Kip3.
We photobleached a linear region along the spindle midzone in cdc15-2–inactivated cells expressing GFP-tubulin and monitored fluorescence recovery (Fig. 3 a). Anaphase spindles photobleached before reaching full length displayed robust recovery of midzone microtubule fluorescence in both control and kip3Δ cells (Fig. 3, Videos 1–4). This fluorescence recovery reflects the contributions from both microtubule dynamics within the midzone as well as antiparallel microtubule sliding, which requires net polymerization of dynamic midzone microtubules. FRAP analysis of full-length spindles in control cells revealed a dramatically different behavior. There was essentially no recovery of midzone microtubule fluorescence after photobleaching (Fig. 3, a–c). Thus, the midzone region switches from a dynamic state to a relatively nondynamic state when spindles reach the proper length and elongation ceases. Strikingly, in the absence of Kip3 this switch failed to occur. In kip3Δ cells, midzone fluorescence recovery remained robust after photobleaching of full-length, fishhook, or hyper-elongated spindles (Fig. 3, a–c). Therefore, the FRAP results reveal that Kip3 mediates a distinct switch from a dynamic to nondynamic midzone region when anaphase spindles reach proper length and stop elongating.
Figure 3.
Kip3 mediates a transition from dynamic to nondynamic spindle midzone when anaphase spindles reach the appropriate length. (a) FRAP analysis of spindle midzone microtubules before (elongating) or after (full length) anaphase spindles reached a length equal to cell length. Spindle midzones in cdc15-2 control cells (rows 1–2) or cells lacking Kip3 (rows 3–4) were photobleached during incubation at 37°C and fluorescence recovery was monitored over time. (b) Normalized midzone fluorescence recovery for representative spindles in the cell types corresponding to the rows in panel a. The average midzone fluorescence recovery rate ± SE for each cell type is plotted in panel c. P < 0.05 for full-length control versus elongating control, and for full-length control versus full-length kip3Δ spindles. For control n = 9 and 13, and for kip3Δ n = 10 and 9 for elongating and full-length spindles, respectively. Bar, 2 µm. See also Videos 1–4.
Overall, in cells lacking Kip3, both the rate of spindle hyper-elongation (Fig. 2 e) and of midzone fluorescence recovery of full-length spindles (Fig. 3 c) were comparable to normal anaphase rates. Together, these results suggest that the mechanisms that drive typical anaphase elongation continue to operate during spindle hyper-elongation in the absence of Kip3.
Kip3 suppresses midzone microtubule dynamics and limits midzone length
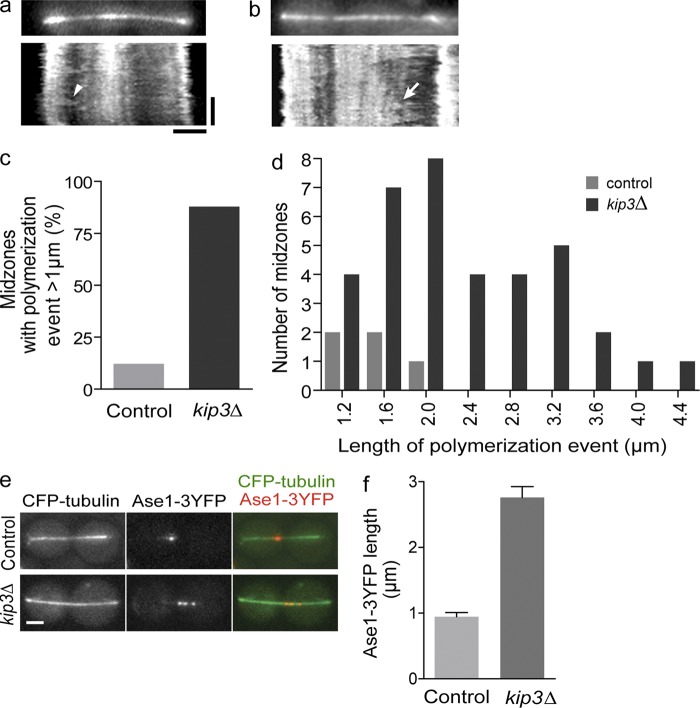
FRAP analysis indicated that Kip3 limits overall midzone dynamics in full-length anaphase spindles, but the experimental setup could not distinguish the contribution of microtubule dynamics from that of antiparallel sliding. Therefore, to determine the effect of Kip3 on spindle microtubule dynamics, independent from that of microtubule sliding, we performed high resolution time-lapse imaging of full-length spindles in cells expressing GFP-tubulin. Kymograph analysis of these spindles allowed us to directly monitor the dynamic behavior of spindle microtubule plus-ends. Microtubules that polymerized out from the spindle poles toward the spindle midzone were frequently observed as diagonal regions of increased fluorescence (Fig. 4 a; and Video 5). In control cells, these microtubules rarely reached the spindle midzone before undergoing catastrophe and depolymerizing rapidly back to the pole. Additionally, microtubules that composed the spindle midzone region, or the sparse microtubule that was observed to grow from the pole across the midzone, rarely experienced notable polymerization events toward the opposite spindle pole.
Figure 4.
Kip3 suppresses spindle microtubule dynamics and limits midzone length during anaphase. (a and b) Late anaphase spindles in control (a) and kip3Δ (b) cells expressing GFP-tubulin (top) were imaged over time to generate kymographs (bottom). In control and kip3Δ cells, microtubules polymerizing out from the spindle poles toward the midzone were observed as diagonal regions of increased fluorescence (arrowhead). In kip3Δ cells, polymerization events were observed extending out of the midzone toward the spindle poles (arrow). (c) Percentage of spindles that displayed microtubule polymerization >1 µm from the midzone toward the spindle pole. For control and kip3Δ cells, n = 41 and 48, respectively. (d) Magnitude of the longest polymerization event for each spindle scored in panel c, P < 0.05. (e) Fluorescence images of Ase1-3YFP localization on late anaphase spindles (CFP-tubulin). (f) Length of Ase1-3YFP region after 1 h anaphase arrest. Average ± SE, P < 0.005, n = 90 from three experiments for each cell type. Bars: (horizontal) 2 µm; (vertical) 120 s.
In contrast to control cells, in kip3Δ cells spindle microtubules extending from the poles often polymerized through the spindle midzone and into the opposite spindle half. Consistent with this, microtubules that extended beyond the midzone frequently underwent significant polymerization toward the opposite spindle pole (Fig. 4 b; and Video 6). When we quantified polymerization events within each spindle, we found that spindle microtubules extending beyond the midzone underwent polymerization events of greater than 1 µm in 80% of kip3Δ cells (Fig. 4 c). By comparison, such polymerization events were observed in only 12% of control spindles. Furthermore, when Kip3 was absent, microtubules polymerized further into the opposite spindle half (Fig. 4 d). When microtubules polymerized more than 1 µm into the opposite spindle half, the maximal length increase was 2.4 ± 0.14 µm in kip3Δ, compared with 1.6 ± 0.14 µm in control cells (P < 0.05). Together, these analyses demonstrate that Kip3 limits the extent of microtubule polymerization beyond the spindle midzone, and suggest that Kip3 prevents spindle hyper-elongation by suppressing polymerization of midzone microtubules.
The majority of forces driving spindle elongation in budding yeast are generated within the spindle midzone (Straight et al., 1998; Schuyler et al., 2003). In the absence of Kip3, spindle length regulation is lost and spindles hyper-elongate. In light of the results from our microtubule dynamics analyses, we hypothesized that the increased polymerization of midzone microtubules in kip3Δ cells would result in an increased region of antiparallel overlap within the spindle midzone, which in turn may drive continued and/or excessive elongation of full-length spindles (Khmelinskii et al., 2009). Ase1, the PRC1/MAP65 homologue, is an antiparallel microtubule cross-linker that localizes to the midzone and is widely used as a midzone marker on anaphase spindles (Schuyler et al., 2003; Janson et al., 2007; Khmelinskii et al., 2007). To test our hypothesis, we visualized Ase1-3YFP on anaphase spindles in the presence or absence of Kip3 (Fig. 4 e). After 1 h of anaphase arrest, the region of the spindle decorated by Ase1-3YFP in kip3Δ cells was ∼200% larger than in control cells (Fig. 4 f). Thus, Kip3 limits the region of antiparallel microtubule overlap within the spindle midzone, likely by suppressing extensive polymerization of midzone microtubules.
Compared with control cells, in kip3Δ cells we observed an increase in spindles with discontinuous localization of Ase1-3YFP. We surmise this may result from local concentration of Ase1-3YFP in areas where antiparallel overlap is repeatedly extended and then shortened due to increased microtubule dynamics (Braun et al., 2011). Alternatively, it may result from cooperative multimerization of bound Ase1 (Kapitein et al., 2008) or altered organization of the overlapped region within the midzone. Overall, these data indicate that increased polymerization of midzone microtubules in the absence of Kip3 results in an increased region of antiparallel overlap within the midzone.
Kip3 is required to suppress the microtubule-polymerizing activity of Stu2
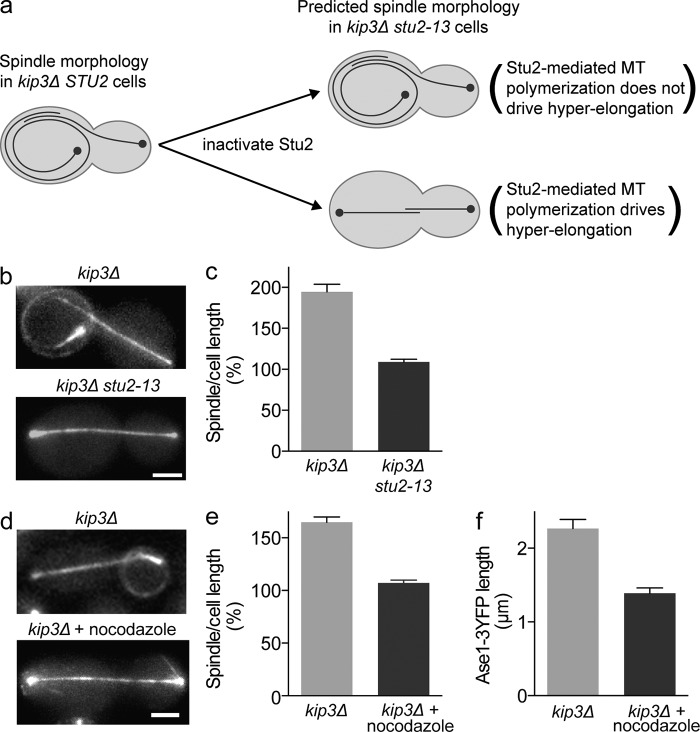
In the absence of Kip3, spindle elongation forces overcome the cortex-imposed resistance by buckling the spindle microtubules, leading to uncontrolled spindle elongation. Our results suggest that Kip3 regulates spindle elongation by limiting midzone microtubule polymerization. Therefore, we hypothesized that Kip3 may be required to oppose factors that promote microtubule polymerization within the spindle. To test this idea, we inactivated the XMAP215/Dis1 family microtubule polymerase, Stu2. We reasoned that if Stu2-mediated microtubule polymerization is a major factor driving spindle hyper-elongation, then the removal of Stu2 should prevent spindle hyper-elongation in kip3Δ cells (Fig. 5 a). Consistent with this, when we inactivated the temperature-sensitive allele stu2-13 in cells lacking Kip3 we no longer observed spindle hyper-elongation (Fig. 5, b and c). Interestingly, in cells lacking both Kip3 and Stu2 activity we found that anaphase spindle length scaled properly with cell size (Fig. 5 c). This result reveals that when microtubule polymerization and depolymerization are adequately balanced the cell boundary can appropriately halt spindle elongation.
Figure 5.
Suppressing microtubule polymerization reduces midzone length and rescues spindle hyper-elongation in the absence of Kip3. (a) Predicted effects of Stu2 inactivation on spindle hyper-elongation. MT, microtubule. (b) Representative spindle morphology in cdc15-2 kip3Δ cells with functional STU2 (top) or inactivated stu2-13 (bottom) during anaphase arrest. (c) Spindle length as a percentage of cell length in anaphase-arrested kip3Δ cells in the presence or absence of active Stu2 (cdc15-2 kip2Δ kip3Δ, 2.5 h at 37°C). (d) Representative GFP-tubulin fluorescence images of kip3Δ cells arrested in anaphase for 1 h, followed by an additional 1.5 h in the absence (top) or presence (bottom) of nocodazole to inhibit microtubule polymerization. (e) Spindle length as a percentage of cell length for cells treated in panel d. (f) Length of Ase1-3YFP region after 2 h anaphase arrest in the presence or absence of nocodazole. Average ± SE: (c) n = 42–45 cells from three experiments; (e) n = 98 cells for each treatment from three experiments; (f) n = 78 and 77 cells for the absence or presence of nocodazole, respectively, from three experiments. P < 0.005 (c, e, and f). Bar, 2 µm.
It was previously shown that Kip3 and Stu2 contribute opposing activities during early spindle elongation (Severin et al., 2001). Spindles failed to elongate in cells lacking Stu2 (Severin et al., 2001). Yet, spindle elongation was restored in the absence of both Stu2 and Kip3 (Severin et al., 2001). Thus, Stu2 is required to drive spindle elongation at the beginning of anaphase, but the functional contribution of Kip3 in this relationship has remained unclear. Kip3 is not required for spindle elongation (Straight et al., 1998). However, in stark contrast, our results demonstrate that once spindles reach full length, it is Kip3 that is required to antagonize Stu2 and properly terminate anaphase spindle elongation.
Blocking microtubule polymerization reduces midzone length and rescues spindle hyper-elongation in cells lacking Kip3
Our data indicate that the mechanism through which Kip3 scales spindle length to cell length is by suppressing the polymerization of midzone microtubules. Thus, we reasoned that in cells lacking Kip3 activity, the direct inhibition of microtubule polymerization in late anaphase would prevent spindle hyper-elongation. To test this, we arrested kip3Δ cells in anaphase until spindles had reached approximately full length, and then continued the arrest in the presence or absence of the drug nocodazole, which inhibits microtubule polymerization. In untreated cells, spindles proceeded to hyper-elongate during the arrest (Fig. 5 d). Strikingly, in the nocodazole-treated cells spindles did not hyper-elongate, rather the spindle length closely matched that of the cell (Fig. 5 e). This result clearly demonstrates that suppression of spindle microtubule polymerization limits anaphase spindle elongation.
Mimicking the microtubule-destabilizing activity of Kip3 with nocodazole treatment rescued control of spindle length in cells lacking Kip3. Therefore, if Kip3 regulates spindle length by restricting midzone size, then nocodazole treatment would be expected to limit the midzone elongation seen in the absence of Kip3. To test this prediction, we measured the midzone length in anaphase-arrested kip3Δ cells expressing ASE1-3YFP in the presence or absence of nocodazole. As predicted, the region decorated by Ase1-3YFP was reduced on average from 2.3 ± 0.1 µm in untreated cells to 1.4 ± 0.1 µm in nocodazole-treated kip3Δ cells (Fig. 5 f). Therefore, we conclude that Kip3, similar to nocodazole treatment, regulates elongation and limits the length of the anaphase spindle by directly suppressing extensive polymerization of midzone microtubules.
The microtubule motility and antiparallel sliding activities of Kip3 are not sufficient to regulate spindle elongation
Taken together, our results demonstrate that Kip3 regulates anaphase spindle length by suppressing polymerization of midzone microtubules. Kip3 can directly destabilize microtubules in vitro (Gupta et al., 2006; Varga et al., 2006). However, it is also a plus end–directed motor (Gupta et al., 2006; Varga et al., 2006), and it was recently shown that Kip3 can cross-link and slide antiparallel microtubules (Su et al., 2013). To molecularly dissect these activities and determine if the microtubule depolymerase activity of Kip3 is required to scale anaphase spindle length, we replaced endogenous Kip3 with Kip3-CC. Kip3-CC is a well-characterized separation of function mutant in which the coiled-coil neck region of Kip3 is replaced with a leucine zipper dimerization motif (Su et al., 2013). Notably, although Kip3-CC remains a highly processive motor with antiparallel sliding activity comparable to Kip3, it is only 10% as effective as Kip3 in its depolymerase activity (Su et al., 2013).
To determine whether the depolymerase activity of Kip3 is required to regulate anaphase spindle length, we subjected cells containing Kip3-CC-EYFP at the endogenous locus to anaphase arrest. As previously reported, Kip3-CC and Kip3 remained similarly localized to anaphase spindles after a shift to the restrictive temperature (Fig. 6 a). In cells containing Kip3, anaphase spindles remained appropriately scaled to cell length. In contrast, however, anaphase spindles in Kip3-CC cells continued to hyper-elongate (Fig. 6 b) similar to spindles in cells lacking Kip3 (Fig. 2). These results clearly demonstrate that the microtubule motility and sliding activities of Kip3 cannot regulate anaphase spindle elongation, and that Kip3 depolymerase activity is essential. Furthermore, they support the conclusion that Kip3 controls anaphase spindle length by directly regulating midzone microtubule polymerization.
Figure 6.
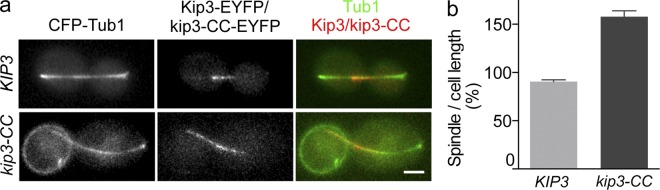
The microtubule motility and sliding activities of Kip3 are not sufficient to regulate spindle length in the absence of microtubule-destabilizing activity. (a) Representative fluorescence images of Kip3-EYFP and kip3-CC-EYFP on spindles (CFP-tubulin) in anaphase-arrested cells (cdc15-2, 2.5 h at 37°C). (b) Spindle length as a percentage of cell length in cells containing Kip3-EYFP or kip3-CC-EYFP after 2.5 h anaphase arrest. Average ± SE, P < 0.005, n = 90 cells from three experiments for each cell type. Bar, 2 µm.
Discussion
We screened mutants known to generate fishhook spindles and identified a novel role for the kinesin-8, Kip3, in regulating spindle elongation. Our data reveal that a Kip3-dependent mechanism operates within the spindle midzone and is required to scale the anaphase spindle length with that of the cell. Together, our results support a model in which Kip3 regulates spindle length by directly suppressing the polymerization of midzone microtubules (Fig. 7). Kip3 limits the extent of microtubule overlap in the midzone, and thus limits the force generated by midzone-associated motor proteins. As a result, elongation forces cannot buckle the spindle microtubules, and thus the cell cortex becomes a significant obstacle that limits the extent of spindle elongation. These findings demonstrate that, when spindle elongation forces are tuned such that the cortical boundary can halt elongation, anaphase spindle length will be directly coupled to the length of the cellular compartment. Such cortical resistance could be encountered directly by the spindle pole, as is likely the case in yeast, or transmitted through the astral microtubule arrays common in higher eukaryotes (Gittes et al., 1993; Dogterom and Yurke, 1997).
Figure 7.
Model for the regulation of anaphase spindle elongation via control of midzone microtubule dynamics by Kip3. Kip3 is required to ensure that anaphase spindle elongation terminates when spindles reach a length equal to cell length (top). Stu2-mediated spindle microtubule polymerization is suppressed by Kip3, which limits antiparallel microtubule overlap in the midzone. As a result, midzone-associated elongation forces will not buckle spindle microtubules, elongation terminates upon resistance from the cell cortex, and spindle length is directly coupled to the length of the cell. In the absence of Kip3 (bottom), midzone length is increased and elongation forces buckle spindle microtubules in response to resistance from the cell cortex. As a result, spindle elongation continues and spindle length exceeds cell length.
During anaphase, Kip3 could regulate midzone microtubules in a constitutive or a length-dependent manner. In the first possibility, Kip3 would constitutively limit midzone microtubule overlap so that elongation forces can overcome cytoplasmic resistance but not buckle the spindle due to opposition from the cell cortex. Consistent with this model, the average size of the ASE1-GFP midzone region did not decrease significantly throughout spindle elongation (Khmelinskii et al., 2007). Alternatively, Kip3 may increasingly suppress midzone microtubule polymerization, and hence spindle elongation, as spindle length is increased. In vitro, Kip3 depolymerized stabilized microtubules in a length-dependent manner (Varga et al., 2006). Consistent with this, FRAP analysis revealed that microtubules are less dynamic in longer spindles relative to those of intermediate length (Fridman et al., 2009). However, when dynein-mediated spindle positioning is defective, yeast cells with mispositioned spindles can arrest in anaphase for hours with spindles approximately half that of a typical full-length spindle (Moore et al., 2009). This observation illustrates that there is not a threshold length at which Kip3 terminates spindle elongation, and that additional mechanisms including cortical resistance are important for controlling spindle elongation.
Preventing the growth of midzone microtubules could inhibit spindle elongation by at least two mechanisms. In the absence of Kip3 we observed increased microtubule polymerization and antiparallel microtubule overlap in the midzone. This would create more binding sites for the kinesin-5 antiparallel sliding motors, Cin8 and Kip1, and may render elongation forces sufficient to buckle the spindle. Therefore, one possibility is that restricting midzone size could limit midzone-associated forces and ensure that spindle elongation cannot exceed the cell boundary. A second mechanism involves the bidirectional motility of Cin8 observed in vitro (Gerson-Gurwitz et al., 2011; Roostalu et al., 2011). High numbers of motors bound to the same microtubule moved collectively toward the plus end, whereas low motor densities traveled toward the minus end (Roostalu et al., 2011). A speculative outcome of this activity is that midzone size may provide feedback for spindle elongation. Increased midzone overlap would increase Cin8 binding and generate plus end–directed sliding and spindle elongation. If sliding outpaced midzone growth, the number of Cin8 motors engaged would decrease until net motility became zero. In this model, if the midzone region became too small, the remaining Cin8 molecules would move toward the minus end and resist further elongation that could compromise spindle stability. Consistent with this idea, Cin8, the Caenorhabditis elegans kinesin-5, BMK-1, and the mammalian kinesin-5, Eg5, were each observed to operate as a “brake” during anaphase B spindle elongation (Saunders et al., 2007; Rozelle et al., 2011; Collins et al., 2014). These two mechanisms are not mutually exclusive and could work together to control spindle elongation while maintaining midzone integrity.
Defining the mechanisms that regulate anaphase spindle length has been challenging because spindle disassembly typically occurs soon after anaphase elongation. However, anaphase spindle elongation is likely regulated in diverse species. In metaphase spindles, the Drosophila kinesin-8, Klp67A, was shown to inhibit spindle microtubules from polymerizing beyond the central overlapped region (Wang et al., 2010). Moreover, knockdown of Klp67A resulted in extra-long spindles during anaphase (Gandhi et al., 2004; Goshima and Vale, 2005; Wang et al., 2010). Thus, it’s likely that kinesin-8 also controls anaphase spindle elongation in Drosophila through similar mechanisms as we find in yeast. Additionally, in monocotyledonous plants, anaphase spindle elongation was observed to terminate in an apparent regulated manner during meiosis (Shamina et al., 2009). In HeLa cells, the kinesin Kif4 negatively regulated midzone microtubule dynamics, and the distance between segregated chromosomes stopped increasing during cytokinesis in a Kif4-dependent manner (Hu et al., 2011). Together, these studies reveal that control of anaphase spindle elongation through the regulation of midzone microtubule dynamics is likely conserved among diverse eukaryotes.
Materials and methods
Strains
Yeast strains and plasmids are described in Table S1. Yeast media and genetic techniques were performed as described previously (Rose et al., 1990). Gene deletions and mutations were introduced by genetic crossing or by fragment-mediated homologous recombination. Unless otherwise specified, yeast strains were of S288C background. Details of strain construction are available upon request.
Microscopy
Cells were grown to log phase in SC complete media (0.67% yeast nitrogen base without amino acids, 2% glucose, and supplemented with amino acids) and placed onto a microscope slide padded with 1% agarose in the same medium. Coverslips were sealed using VALAP. Fluorescence imaging was performed using a CCD camera (CoolSNAP HQ2; Photometrics) mounted on a microscope (AxioImager M2; Carl Zeiss) with a piezoelectric-driven Z-stage, 63× 1.4 NA Plan Apochromat objective, and Semrock filters, driven by SlideBook software (Intelligent Imaging Innovations, Inc.). During short- and long-term imaging of live cells at restrictive temperature, specimens were maintained at 36–37°C using an air-stream incubator (400 ASI; Nevtek, Inc.). Unless otherwise stated, cells were imaged at room temperature.
Spindle size and elongation rates
For population experiments tracking spindle length over time, log-phase cultures were incubated at restrictive temperature and imaged for GFP fluorescence at 37°C with 10–14 Z-plane images spaced 0.5 µm apart. To determine spindle elongation and hyper-elongation rates, similar images were obtained every 60 s for at least 60 min. For nocodazole treatment, log-phase cultures were incubated at 37°C for 1 h and then the SC media was supplemented with 10% pre-warmed YPD media (1% yeast extract, 2% peptone, 2% glucose) to aid nocodazole solubility. Next, the culture was split and treated with nocodazole (Sigma-Aldrich) at a final concentration of 25 µg/ml and 1% DMSO or with 1% DMSO alone. Cultures were incubated another 1.5 h at 37°C and then imaged live at 37°C. To determine spindle length after stu2-13 inactivation, cultures were incubated at restrictive temperature (37°C) for 2.5 h and spindles were imaged by GFP-tubulin fluorescence as above. Spindle lengths were measured using SlideBook software by determining the length of a line that tracked the spindle from pole to pole, taking into account distance in the Z-plane. Elongation rates were determined by plotting the spindle length against time throughout the video.
FRAP of spindle midzones
For midzone FRAP experiments cells were imaged, in the medium described above, for 300 ms in single Z-planes at 5-s intervals using an EM-CCD camera (Cascade 512B; Photometrics) mounted on a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a spinning-disk confocal attachment (CSU10; Yokogawa Corporation of America) with a 63× 1.4 NA Plan Apochromat objective. The microscope was controlled using MetaMorph software (Molecular Devices). For photobleaching, a galvanometer-steerable 440-nm dye laser was used (MicroPoint; Photonics Instruments, Inc.). Temperature was controlled using an air-stream incubator (400 ASI; Nevtek, Inc.). Unless otherwise stated, cells were imaged at room temperature.
To determine FRAP, a region was defined that encompassed the spindle midzone and the area and integrated fluorescence intensity measured for each time-lapse image. Time points in which one spindle pole and/or the spindle midzone were not in focus, or astral microtubules overlapped with the midzone region, were omitted from the analysis. The integrated cytoplasmic background intensity was subtracted from the integrated midzone intensity, and the difference corrected for photobleaching during image acquisition. The post-bleach midzone intensity values were normalized to the pre-bleach value and averaged across three consecutive time points in a sliding window. Because the recovered fluorescence represents both dynamic instability and net polymerization, and not simply a direct exchange of tubulin subunits, the increase in midzone fluorescence, before reaching a plateau, was analyzed by linear regression and reported as percent recovery over time.
Spindle microtubule dynamics
For imaging of midzone microtubule dynamics, 7–9 Z-plane images spaced 0.4 µm apart were collected every 10 s. For analysis, only full-length or greater spindles were used. Maximum intensity Z-projections were prepared and used to generate kymographs of fluorescence signal along the spindle using ImageJ software (National Institutes of Health). The projection images of fishhook spindles were processed with the “Straighten” ImageJ plugin tool before generating the kymograph. Kymographs of spindles from control and kip3Δ cells were randomized and scored by blind analysis. Each kymograph was examined for the presence of microtubule polymerization across the midzone toward the opposite spindle pole. If present, the length of the longest event in each kymograph was determined using ImageJ.
Ase1-3YFP localization
Log-phase cultures were synchronized using hydroxyurea (Sigma-Aldrich) added to a final concentration of 0.2 M for 1.5 h. Hydroxyurea was then rinsed out with a series of three washes with SC media, and the resuspended cultures were incubated at 37°C for 1 h. For nocodazole treatment, log-phase cultures in SC were incubated at 37°C for 1 h before supplementation with 10% pre-warmed YPD media. The culture was split and treated with either 25 µg/ml nocodazole and 1% DMSO, or 1% DMSO alone. Cultures were incubated another 1.5 h at 37°C and then imaged live at 37°C. Imaging conditions were maintained at 300-ms exposure for CFP, and 500-ms exposure for YFP fluorescence over 14 Z-planes spaced 0.5 µm apart. For analysis, the intensity of all images was adjusted equally using SlideBook.
Screening of fishhook spindle mutants
Log-phase cultures were incubated at 37°C for 2.5 h. These cultures were then imaged live at 37°C for GFP-tubulin fluorescence over 14 Z-planes spaced 0.5 µm apart. Experiments were conducted at least three times and lasso spindles were visually scored as those that exceeded cell length to the extent that the spindle microtubule polymer wrapped along the cell cortex and doubled back on itself. To ensure that the function of Kip3 in controlling spindle length was not specific to a particular yeast genetic background, we confirmed the results we observed in the S288C genetic background in cells from the W303 background (Fig. S1, d and e). The fishhook mutants mcm22Δ and hnt3Δ (Vizeacoumar et al., 2010) were not tested, as we were unable after multiple attempts to generate strains combining the mutations with the cdc15-2 allele.
Kip3-EYFP and kip3-CC-EYFP localization
Log-phase cultures were incubated at 37°C for 2.5 h before being imaged live at 37°C. 14 Z-planes spaced 0.5 µm apart were captured for CFP and YFP fluorescence with 300-ms and 400-ms exposures, respectively. For analysis, the intensity of all images was adjusted equally using SlideBook.
Statistical analysis
P-values were determined by unpaired Student’s t test using Prism (GraphPad Software). Averages are reported ± SE.
Online supplemental material
Fig. S1 demonstrates that deletion of KIP2 does not affect spindle length regulation or morphology, and that spindles hyper-elongate in the W303 genetic background. Table S1 lists all yeast strains and plasmids used in this study. There are six videos (Videos 1–6) that correspond, respectively, to Fig. 3 a (first row), Fig. 3 a (second row), Fig. 3 a (third row), Fig. 3 a (fourth row), Fig. 4 a, and Fig. 4 b. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201312039/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201312039.dv.
Supplementary Material
Acknowledgments
We thank Y. Fukuda, T. Hyman, A. Luchniak, and E. Murphy for valuable discussions; K. Bloom, M. Glotzer, D. Laporte, S. Rizk, I. Sagot, and Z. Storchova for helpful feedback on this manuscript; and M. Guerguis for graphical design of Fig. 7. We thank T. Huffaker, B. Lavoie, and D. Pellman for reagents.
R.S. Rizk was supported by a National Institutes of Health postdoctoral training fellowship (T32 HL094282) and an American Heart Association postdoctoral fellowship. K. DiScipio received a University of Chicago – Biological Sciences Collegiate Division undergraduate fellowship. This work was supported by a National Institutes of Health grant (R01GM094313) and funding from the Illinois Division of the American Cancer Society (214937) to M.L. Gupta.
The authors declare no competing financial interests.
References
- Braun M., Lansky Z., Fink G., Ruhnow F., Diez S., Janson M.E. 2011. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat. Cell Biol. 13:1259–1264 10.1038/ncb2323 [DOI] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M.L., Jr, Hoyt M.A., Pellman D. 2004. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 6:815–829 10.1016/j.devcel.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Collins E., Mann B.J., Wadsworth P. 2014. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken). 71:136–144 10.1002/cm.21158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F.R., Hoyt M.A. 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138:1041–1053 10.1083/jcb.138.5.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti J., Hartwell L.H. 1971. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp. Cell Res. 67:389–401 10.1016/0014-4827(71)90424-1 [DOI] [PubMed] [Google Scholar]
- Dogterom M., Yurke B. 1997. Measurement of the force-velocity relation for growing microtubules. Science. 278:856–860 10.1126/science.278.5339.856 [DOI] [PubMed] [Google Scholar]
- Du Y., English C.A., Ohi R. 2010. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 20:374–380 10.1016/j.cub.2009.12.049 [DOI] [PubMed] [Google Scholar]
- Erent M., Drummond D.R., Cross R.A. 2012. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS ONE. 7:e30738 10.1371/journal.pone.0030738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa J.P., Gerlich D.W. 2012. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14:440–447 10.1038/ncb2482 [DOI] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M., Gheber L. 2009. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10:387–393 10.1038/embor.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R., Bonaccorsi S., Wentworth D., Doxsey S., Gatti M., Pereira A. 2004. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol. Biol. Cell. 15:121–131 10.1091/mbc.E03-05-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Koonrugsa N., Toda T. 2002. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12:610–621 10.1016/S0960-9822(02)00761-3 [DOI] [PubMed] [Google Scholar]
- Gardner M.K., Zanic M., Gell C., Bormuth V., Howard J. 2011. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 147:1092–1103 10.1016/j.cell.2011.10.037 [DOI] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D.R., Schmidt C.F., Gheber L. 2011. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30:4942–4954 10.1038/emboj.2011.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F., Mickey B., Nettleton J., Howard J. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923–934 10.1083/jcb.120.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Scholey J.M. 2010. Control of mitotic spindle length. Annu. Rev. Cell Dev. Biol. 26:21–57 10.1146/annurev-cellbio-100109-104006 [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907 10.1091/mbc.E05-02-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L., Jr, Carvalho P., Roof D.M., Pellman D. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Hara Y., Kimura A. 2009. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr. Biol. 19:1549–1554 10.1016/j.cub.2009.07.050 [DOI] [PubMed] [Google Scholar]
- Hu C.-K., Coughlin M., Field C.M., Mitchison T.J. 2011. KIF4 regulates midzone length during cytokinesis. Curr. Biol. 21:815–824 10.1016/j.cub.2011.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson M.E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F.J., Tran P.T. 2007. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 128:357–368 10.1016/j.cell.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Janson M.E., van den Wildenberg S.M.J.L., Hoogenraad C.C., Schmidt C.F., Peterman E.J.G. 2008. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr. Biol. 18:1713–1717 10.1016/j.cub.2008.09.046 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 17:244–256 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Kotadia S., Montembault E., Sullivan W., Royou A. 2012. Cell elongation is an adaptive response for clearing long chromatid arms from the cleavage plane. J. Cell Biol. 199:745–753 10.1083/jcb.201208041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C., Winey M. 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 9:29–46 10.1091/mbc.9.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., Salmon E.D. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr M.I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G., Mayer T.U. 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- McNally F.J. 2013. Mechanisms of spindle positioning. J. Cell Biol. 200:131–140 10.1083/jcb.201210007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Heller K.K., Frisèn L., Wallack D.L., Loayza D., Gammie A.E., Rose M.D. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell. 9:2051–2068 10.1091/mbc.9.8.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Magidson V., Khodjakov A., Cooper J.A. 2009. The spindle position checkpoint requires positional feedback from cytoplasmic microtubules. Curr. Biol. 19:2026–2030 10.1016/j.cub.2009.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J., Schiebel E., Khmelinskii A. 2010. Cell cycle control of spindle elongation. Cell Cycle. 9:1084–1090 10.4161/cc.9.6.11017 [DOI] [PubMed] [Google Scholar]
- Roostalu J., Hentrich C., Bieling P., Telley I.A., Schiebel E., Surrey T. 2011. Directional switching of the kinesin Cin8 through motor coupling. Science. 332:94–99 10.1126/science.1199945 [DOI] [PubMed] [Google Scholar]
- Rose M.D., Winston F., Hieter P. 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Rozelle D.K., Hansen S.D., Kaplan K.B. 2011. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 193:285–294 10.1083/jcb.201011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A.M., Powers J., Strome S., Saxton W.M. 2007. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 17:R453–R454 10.1016/j.cub.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., Pellman D. 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160:517–528 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. 2001. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442 10.1083/jcb.153.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamina N.V., Zharkov N.A., Omelyanchuk L.V. 2009. Feed-back regulation of successive meiotic cytokinesis. Cell Biol. Int. 33:393–401 10.1016/j.cellbi.2009.01.011 [DOI] [PubMed] [Google Scholar]
- Storchová Z., Breneman A., Cande J., Dunn J., Burbank K., O’Toole E., Pellman D. 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature. 443:541–547 10.1038/nature05178 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. 1998. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143:687–694 10.1083/jcb.143.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., Arellano-Santoyo H., Portran D., Gaillard J., Vantard M., Thery M., Pellman D. 2013. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 15:948–957 10.1038/ncb2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer C., Brunner D., Dogterom M. 2009. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol. Syst. Biol. 5:250 10.1038/msb.2009.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth A., Masuda H., Dhut S., Toda T. 2008. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol. Biol. Cell. 19:5104–5115 10.1091/mbc.E08-02-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- Vizeacoumar F.J., van Dyk N., S Vizeacoumar F., Cheung V., Li J., Sydorskyy Y., Case N., Li Z., Datti A., Nislow C., et al. 2010. Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J. Cell Biol. 188:69–81 10.1083/jcb.200909013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Brust-Mascher I., Cheerambathur D., Scholey J.M. 2010. Coupling between microtubule sliding, plus-end growth and spindle length revealed by kinesin-8 depletion. Cytoskeleton (Hoboken). 67:715–728 10.1002/cm.20482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., Troxell C.L., McIntosh J.R. 2001. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol. Biol. Cell. 12:3919–3932 10.1091/mbc.12.12.3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West R.R., Malmstrom T., McIntosh J.R. 2002. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J. Cell Sci. 115:931–940 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2010. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 191:795–808 10.1083/jcb.201006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhu C., Chen H., Li L., Guo L., Jiang W., Lu S.H. 2010. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 31:1676–1684 10.1093/carcin/bgq134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.