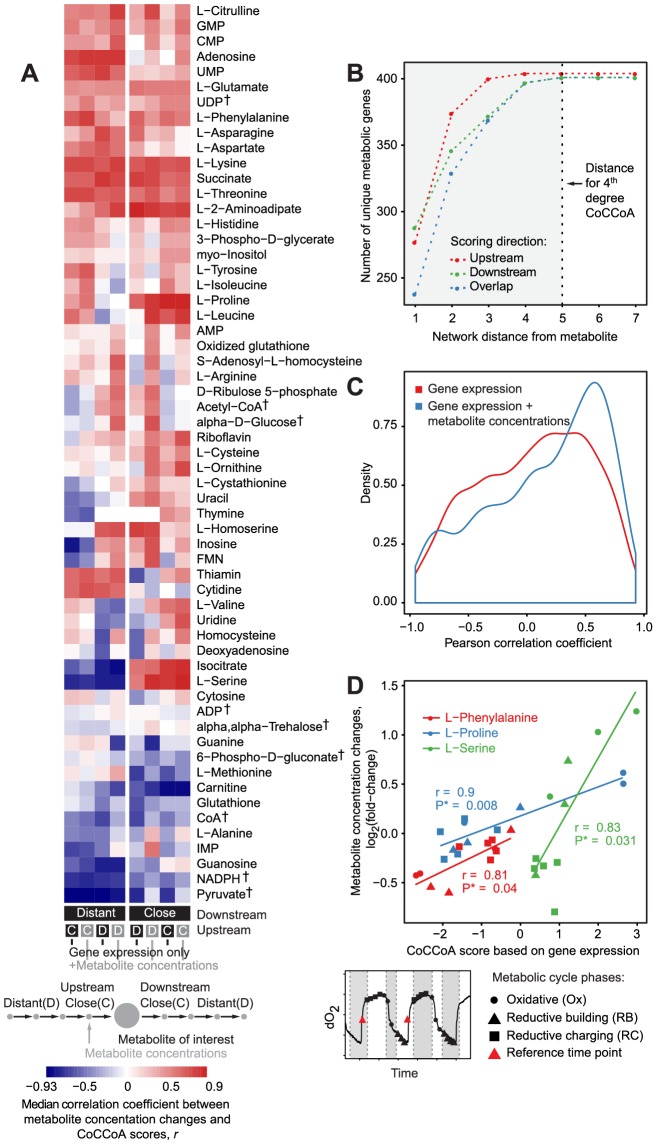
Figure 4. CoCCoA models link the changes in gene expression and metabolite concentrations during yeast metabolic cycle.
A) Pearson correlation coefficients comparing the experimentally observed metabolite concentration changes and CoCCoA model scores. For a given metabolite, 16 different CoCCoA scores were computed, accounting for a network distance of up to 4, both upstream and downstream of the metabolite in question. Scores were divided into two sets: close (distance ≤2) and distant (distance >2). Shown values are the median correlation coefficients in each set. B) CoCCoA models account for a large fraction of genes in the metabolic network at relatively short distances. C) Correlations between the experimentally observed metabolite concentration changes and CoCCoA model scores improve with the inclusion of the concentrations of the precursors and/or products of the metabolite in question. D) Example correlations between the CoCCoA model scores and the metabolite concentration changes. Different shapes of data points mark different stages of the yeast metabolic cycle. † - Metabolites with previous evidence for post-translational regulation of at least one of their neighboring enzymes [34].