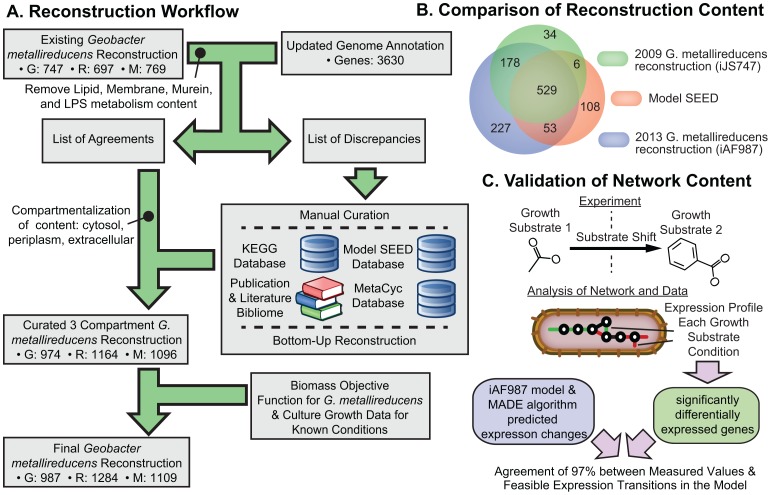
Figure 1. The workflow developed to generate iAF987 along with comparison and validation of its content.
A) The workflow detailing the reconstruction process of the G. metallireducens metabolic network. The reconstruction process was initiated by comparing the updated genome annotation for G. metallireducens to the existing reconstruction to create a list of discrepancies that was manually reviewed and curated. Content that was in agreement with the updated annotation and reconstruction was used to generate a draft set of intracellular reactions. Lipid, membrane, murein, and LPS content were removed from this list as a periplasm compartment was added to the reconstruction. Manual curation [9] was aided by the KEGG [46], ModelSEED [20], and MetaCyc [35] databases. Further, numerous publications and literature sources (i.e., the “bibliome”) were used to refine the network content. The manual review process resulted in a draft reconstruction that was used in conjunction with a formulated biomass objective function in simulations to validate the content of the reconstruction and generate a final version. Some figure images adapted from [47]. B) Venn Diagram showing the comparative analysis of gene content included in different versions of a G. metallireducens reconstruction. C) A schematic of the validation of network content with transcriptomics data for a shift from acetate growth conditions to benzoate with the MADE computational algorithm.