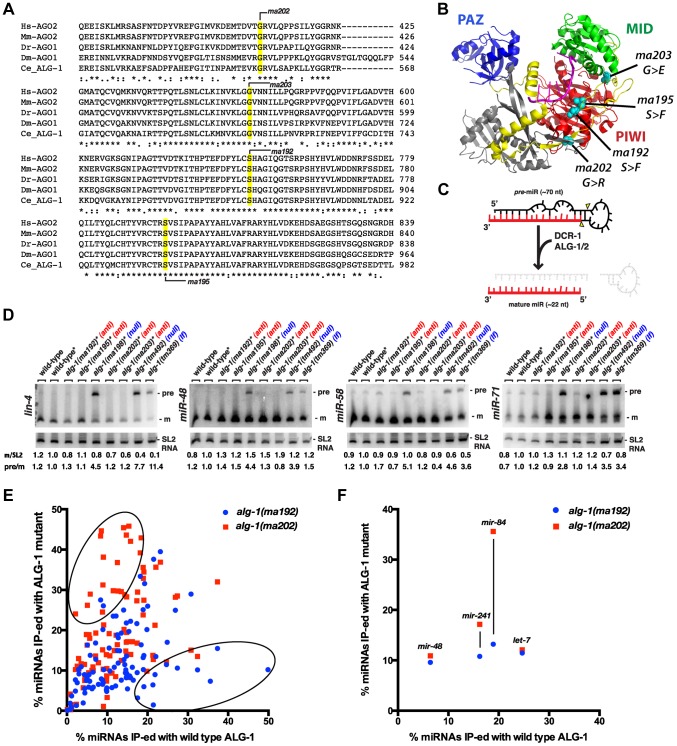
Figure 8. alg-1(anti) alleles affect conserved amino acids within the ALG-1 protein, do not affect pre-microRNA processing, and associate with microRNAs to levels comparable to wild type.
(A) Alignments of Argonaute sequences from multiple species shows conservation of the amino acids affected by alg-1(anti) mutations. (B) Locations of conserved amino acids affected by alg-1(anti) mutations are mapped onto the crystal structure of hAGO2 (PDB ID 4EI1 [51]) using PYMOL. PAZ domain colored in blue, Linker 2 in yellow, MID domain in green, and PIWI domain in red. Amino acids affected by each mutation are highlighted in teal. (C) A schematic showing precursor microRNA processing into the mature guide microRNA. (D) Northern blot analysis of total RNA extracted from mixed population wild type and alg-1 mutant animals using probes to specific microRNAs. Only alg-1(0) null and alg-1(tm369) loss of function mutants accumulate the precursor species of microRNAs. *Strains carry lin-31, col-19::gfp in the background. m/SL2, ratio of mature microRNA to SL2 loading control normalized to wild type lin-31; col-19::gfp. pre/m, ratio of precursor to mature microRNA normalized to wild type lin-31; col-19::gfp. (E) Scatterplot comparing the efficiency with which microRNAs co- immunoprecipitated wild type (X-axis) or mutant (Y-axis) ALG-1. microRNA extracted from ALG-1 immunoprecipitations was quantified using Taqman qRT-PCR. microRNA abundance in each IP was normalized to a synthetic spike-in, and also to the amount of microRNA in the starting material and to the amount of ALG-1 immunoprecipitated (IP-ed). The graph shows average % IP-ed from 3 biological replicates. (F) Subset of data in (E), showing % of let-7-Family microRNAs IP-ed with ALG-1. All strains in (E, F) carry lin-31(lf) and col-19::gfp in the background. The lin-31 mutation is present in order to (non-heterochronically) suppress alg-1(anti) vulval bursting phenotypes.