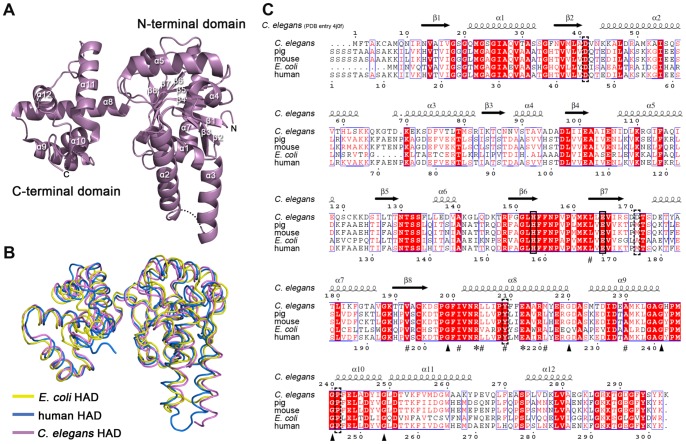
Figure 2. Overall structure of cHAD and protein primary structure comparison between cHAD and its homologues.
(A) Ribbon diagram of cHAD crystal structure (PDB entry 4J0F, this work) colored in purple. The loop between helix α2 and α3 with weak electron density is indicated by dotted line. (B) Tertiary structure alignment among 3-hydroxyacyl-CoA dehydrogenases from C. elegans (purple, PDB entry 4J0F), human (blue, 3HAD) and E. coli (yellow, 3MOG). The region from residue 286 to 475 in E. coli HAD is not shown. (C) Sequence alignment among 3-hydroxyacyl-CoA dehydrogenases from Caenorhabditis elegans (C. elegans, GenBank accession No. CAA80153.1), Homo sapiens (human, GenBank accession No. CAA65528.1), Sus scrofa (pig, GenBank accession No. AAD20939.1), Mus musculus (mouse, GenBank accession No. BAA06122) and Escherichia coli (E. coli, GenBank accession No.NP_415913.1, residues 1–286). The transit peptide sequence was excluded from human, pig and mouse HAD sequences. The secondary structures are corresponding to cHAD. The catalytic His-Glu pair is boxed in black. The disease related point mutations in Table 1 are boxed with dashed line. The conserved glycine residues flanking C-terminal domain helices are indicated by triangles. The conserved Arg and Glu forming salt-bridge on dimerization interface are indicated by asterisks; and the conserved hydrophobic residues on the dimerization interface are indicated by “#”. The labels of secondary structure are corresponding to those in (A).