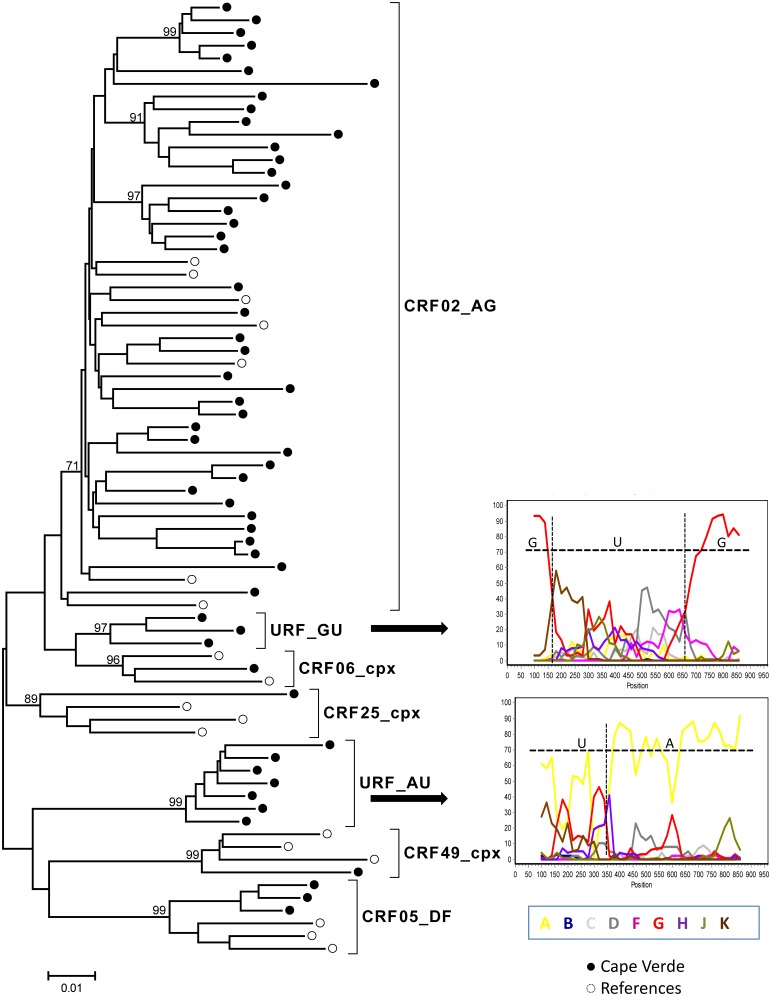
Figure 2. Phylogenetic tree analysis of the pol region of HIV-1 CRF and major URF samples from Cape Verde.
The phylogenetic inferences were performed by the Neighbor-Joining algorithm under the Kimura-2 parameter nucleotide substitution model using the MEGA v5.0 package. The scale represents the number of substitutions per site. Cape Verde and reference sequences are represented, respectively as black and white circles. Bootscan analyses of HIV-1 major URF samples are displayed. Recombinant profiles were inferred using a sliding window of 200 bp, steps of 20 bp and the Kimura-2 parameters model using SimPlot 3.5.1 software. Reference samples corresponding to the major HIV-1 subtypes are indicated by different colors.