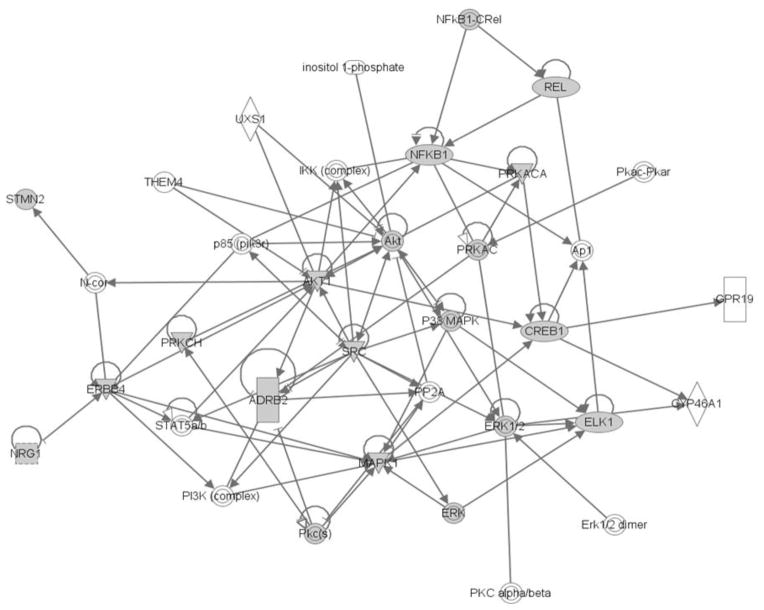
Fig. 4.
NRG1 signalling and neurite formation mediators. Highlighted in grey are molecules within the NRG1-ERBB4 signalling pathway found to be altered in expression (PRKCH and SRC) or phosphorylation (NFkB1, REL, CREB1, ELK1, STMN2 and ADRB2), or kinases directly phosphorylating these molecules (AKT, ERK, MAPK). Other molecules not highlighted in grey are also part of the pathway/network but not found to be altered in our study. Triangles represent kinases; rectangles represent ligand-gated nuclear receptor; concentric circles represent enzyme families; ellipse represents transcription regulators. Figure was generated using IPA pathway analysis software. This software uses a Right tailed Fisher’s exact test to calculate p values of identified pathways, based on the number of significant molecules in the pathway and the fold change of each molecule.