Abstract
The regulation of transcription factor function in response to neuronal activity is important for development and function of the nervous system. The transcription factor Sp4 regulates the developmental patterning of dendrites, contributes to complex processes including learning and memory, and has been linked to psychiatric disorders such as schizophrenia and bipolar disorder. Despite its many roles in the nervous system, the molecular mechanisms regulating Sp4 activity are poorly understood. Here, we report a site of phosphorylation on Sp4 at serine 770 that is decreased in response to membrane depolarization. Inhibition of the voltage-dependent NMDA receptor increased Sp4 phosphorylation. Conversely, stimulation with NMDA reduced the levels of Sp4 phosphorylation, and this was dependent on the protein phosphatase 1/2A (PP1/PP2A). A phospho-mimetic substitution at S770 impaired the Sp4-dependent maturation of cerebellar granule neuron primary dendrites, whereas a non-phosphorylatable Sp4 mutant behaved like wild-type. These data reveal that transcription factor Sp4 is regulated by NMDA receptor-dependent activation of a PP1/PP2A signaling pathway. Our findings also suggest that the regulated control of Sp4 activity is an important mechanism governing the developmental patterning of dendrites.
Keywords: transcription factor Sp4, NMDA receptor, dendrite, phosphorylation, PP1/PP2A, cerebellar granule neurons
Introduction
The regulated activity of transcription factors plays crucial roles in the development and maintenance of connections in the nervous system. Neuronal activity, in the form of membrane depolarization, is known to influence transcription factor activity (Greer & Greenberg 2008). Membrane depolarization activates voltage-gated calcium channels such as the NMDA receptor to initiate signaling cascades, including the activation of downstream kinases and phosphatases, to alter transcription factor function (Cohen & Greenberg 2008, Hardingham 2009, Paoletti et al. 2013, Morishita et al. 2001, Genoux et al. 2002). Thus, many of the profound effects of the NMDA receptor on neuronal development, viability, and plasticity, are mediated, in part, through the regulated post-translational modification of transcription factors.
Sp4 is a zinc-finger transcription factor that is highly expressed in neurons. (Mao et al. 2007) Alterations at the Sp4 gene locus have been linked to psychiatric disorders, including bipolar disorder, major depressive disorder, and schizophrenia (Shi et al. 2011, Shyn et al. 2011, Zhou et al. 2009, Tam et al. 2010). Reduced levels of the Sp4 protein have been directly observed in the cerebellum and prefrontal cortex of bipolar disorder subjects and Sp4 levels in the cerebellum are inversely correlated with severe negative symptoms in schizophrenia (Pinacho et al. 2011, Pinacho et al. 2013). Mice with reduced Sp4 expression displayed deficits in learning and memory and impaired prepulse inhibition, a suggested endophenotype for schizophrenia and other psychiatric disorders (Zhou et al. 2005). Consistent with observed memory deficits, Sp4 hypomorphs exhibited decreased long-term potentiation in hippocampal slice recordings (Zhou et al. 2010).
Sp4 activity is likely to be highly dependent upon the cellular and developmental contexts of its expression. In dentate granule neurons of the hippocampus, Sp4 promotes dendrite outgrowth and branching (Zhou et al. 2007). We have previously shown that in developing cerebellar granule (CG) neurons Sp4 is required for dendritic morphogenesis by limiting dendrite branching and promoting the elimination of excess primary dendrites (Ramos et al. 2007, Ramos et al. 2009). The maturation of CG neuron dendrites is concomitant with the arrival of excitatory mossy fibers, and this process is regulated in vitro by membrane depolarization. These observations suggested that depolarization regulates Sp4 activity, and, indeed, depolarization enhances the stability of the Sp4 protein (Pinacho et al. 2011). The specific pathways that regulate the stability and activity of the Sp4 protein in response to extracellular signals, however, are unknown.
Here, we identify a site of phosphorylation on Sp4 at S770 that is reduced in response to membrane depolarization. We provide evidence that the NMDA receptor dependent activation of a PP1/PP2A signaling pathway reduces Sp4 phosphorylation at S770. Inhibition of the NMDA receptor increased Sp4 S770 phosphorylation while having no effect on the levels of the protein, indicating that S770 phosphorylation and degradation are separable processes. A non-phosphorylatable mutant of Sp4 promoted CG neuron maturation while a phospho-mimetic Sp4 mutant impaired this function, suggesting that the phosphorylation state of Sp4 S770 influences the dendritic maturation of CG neurons. These data describe Sp4 as a transcription factor regulated downstream of NMDA receptor activation, revealing new mechanisms by which neuronal activity informs the gene expression programs of the nervous system.
Materials and Methods
Materials
Nimodipine, 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), MK-801, DL-2-Amino-5-phosphonopentanoic acid (APV), Cyclosporin A, and NMDA were obtained from Sigma. FK-506 was obtained from VWR. Calyculin A was obtained from Cell Signaling Technologies. Okadaic acid was obtained from Millipore. The lambda protein phosphatase was obtained from New England Biolabs and was used according to the manufacturer’s instructions.
Cell culture and treatments
Cerebellar granule neurons were obtained from P6 rats (Charles River Laboratories) and cultured in 25mM KCl as previously described (Bilimoria & Bonni 2008). Cortical neuron cultures were prepared from P0 rats as previously described (Brandon et al. 1999). All protocols regarding the use of animals were approved by the Committee for the Humane Use of Animals at Tufts University School of Medicine. At DIV 4–5, cells were treated for one hour with fresh media containing 25mM KCl (unless otherwise indicated) and inhibitors at the concentrations indicated in the text. Cells were stimulated with NMDA for one hour in magnesium free Locke’s solution without tetrodotoxin as described (Sato et al. 2001). Neuro2A and 293T cells were cultured in DMEM supplemented with 10% Fetalclone (Hyclone).
Plasmids and transfections
Short hairpin RNAs and FLAG-Sp4 were previously described (Ramos et al. 2007). RNAi-resistant FLAG-Sp4 was generated by introducing silent mutations at bp1626, 1629, and 1632. S770 point mutants were generated using site-directed mutagenesis. Deletion mutants were generated by in-frame deletion of FLAG-Sp4 at the amino acids indicated.
For Western blot analysis, cerebellar granule neurons were transfected by nucleofection at DIV0 as described with 20–30% transfection efficiency (Kang et al. 2009). For morphometric analysis, neurons were transfected at DIV 2 using the calcium phosphate method essentially as described with 1–2% transfection efficiency (Konishi et al. 2002). Neuro2A and 293T cells were transfected using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions with 90% transfection efficiency.
Western blotting
Cells were lysed in RIPA buffer supplemented with protease inhibitors (Roche) and phosphatase inhibitors (Santa Cruz). Equal amounts of protein were separated, transferred to nitrocellulose membranes, and incubated with the indicated primary antibody: rabbit polyclonal to Sp4 (Santa Cruz); mouse monoclonal to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Millipore); mouse monoclonal to FLAG (Sigma); chicken polyclonal to GFP (Life Technologies); and rabbit polyclonal to phospho-Sp4 S770 (pSp4). Blots were incubated in horseradish peroxidase conjugated secondary antibodies (Santa Cruz) and linear-range exposures were quantified by densitometry using Fiji software.
Sp4 purification and mass spectrometry
Sp4 was purified from nuclear extracts from P6 rat cerebellum using an Sp4 antibody, digested with trypsin:chymotrypsin, and analyzed by liquid chromatography tandem mass spectrometry (LC/MS/MS) at the Taplin mass spectrometry facility (Harvard Medical School, Boston, MA).
Generation of the phospho-specific antibody
The rabbit antibody to phosphorylated Sp4 at serine 770 was generated against the synthetic phospho-peptide: [CKKAAISQDpSNPATPN] (Covance). Rabbit serum was purified by two-step affinity chromatography against the unmodified and phosphorylated peptide, and validated as indicated in the text.
Immunohistochemistry and tissue processing
Adult rat brains were cut sagittally on a cryostat at 10 microns, mounted, and fixed in 4% paraformaldehyde. After blocking, tissue was incubated overnight at 4°C with Sp4 pSp4, or pSp4 plus 40μg/mL phosphopeptide followed by a biotin conjugated rabbit secondary. Protein was visualized using the DAB reagent kit (Vector laboratories).
Morphometric analysis of dendrites
DIV 5 neurons were washed in phosphate buffered saline, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 5% normal goat serum. Cells were incubated overnight at 4°C with GFP antibody, followed by fluorescent conjugated secondary antibodies (Life Technologies). Dendrites were counted under 40× magnification and statistical analysis was performed as described. Approximately 25–30 neurons were counted per condition and the experiment was repeated three times.
Statistics
All results represent the mean ± SEM from at least 3 independent experiments. Unpaired t-tests or ANOVA followed by Bonferonni’s post hoc test for multiple comparisons were performed where indicated.
Results
Phosphorylation of Sp4 at S770 is regulated by membrane depolarization
In order to study the signal-dependent regulation of Sp4, we treated CG neurons with media containing depolarizing (25mM) or non-depolarizing (resting; 5mM) concentrations of extracellular KCl and examined Sp4 by immunoblot. The transcription factor Sp4 protein is rapidly degraded when CG neurons are cultured in resting conditions ((Pinacho et al. 2011) and Fig. 4B). Compared to depolarizing conditions, a reduction in Sp4 protein mobility was observed in protein extracts from cells cultured in resting conditions (Fig. 1, compare lanes 1 and 3). Phosphatase treatment of the extracts increased Sp4 protein mobility in both conditions (Fig. 1, lanes 2 and 4). These results indicate that Sp4 is a phospho-protein in depolarizing conditions, and is further phosphorylated in resting conditions.
Figure 4. NMDA receptor signaling reduces the levels of phospho-Sp4 S770.
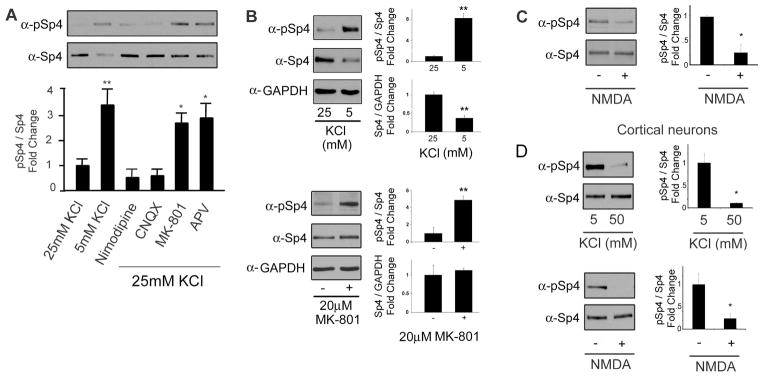
A. CG neurons were treated with the indicated inhibitors and/or KCl concentrations for one hour and protein extracts were analyzed for the expression of phospho-Sp4 S770 (pSp4) and total Sp4 by Western blot. Top – representative immunoblot. Bottom – quantification of the ratio of pSp4 relative to total Sp4 and normalized to the 25mM KCl condition. N=3, *p<0.05, **p<0.01, ANOVA (F=10.85; p<0.0001). B. CG neurons were treated with 25mM or 5mM KCl (Top) or 20μM MK-801 (Bottom) for one hour and protein extracts were analyzed by Western blot. For KCl: N=6, unpaired T-test: pSp4/Sp4 (p<0.0001), Sp4/GAPDH (p<0.0001); For MK-801: N=3, unpaired T-test: pSp4/Sp4 (p=0.0092), Sp4/GAPDH (p=0.67). C. CG neurons were stimulated with or without 100μM NMDA for one hour in magnesium-free Locke’s solution and Sp4 phosphorylation at S770 was analyzed as described in (A) and normalized to the untreated condition. N=7, unpaired T-test (p=0.0255). D. Cortical neurons were depolarized with 50mM KCl (Top) or treated with 100μM NMDA (Bottom) and the levels of Sp4 phosphorylation at S770 relative to total Sp4 were determined by Western blot. N=3, unpaired T-test: KCl (p=0.0120), NMDA (p=0.0474).
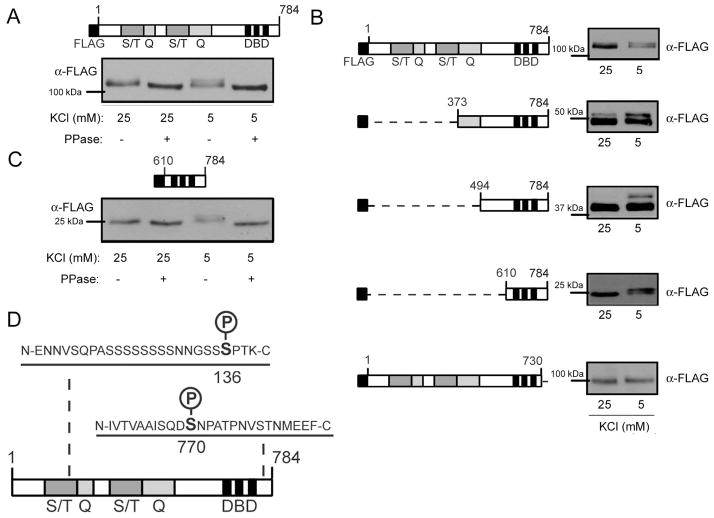
Figure 1. Sp4 is phosphorylated in response to resting membrane potential.
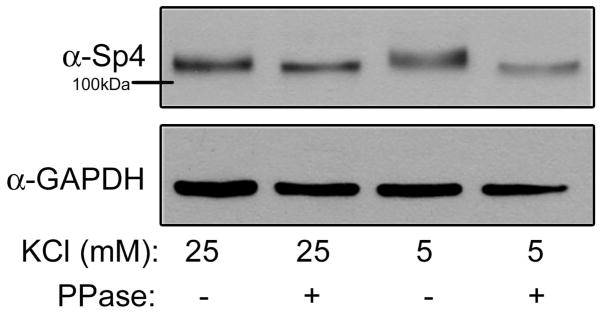
CG neurons were exposed to 25mM (depolarizing) or 5mM (resting) KCl for one hour. Protein extracts were treated with or without a protein phosphatase (PPase), separated by 6% PAGE or 10% PAGE and analyzed by Western blot using antibodies recognizing Sp4 and GAPDH as a loading control. Shown is a representative immunoblot of an experiment performed 5 times.
We employed multiple approaches to identify the Sp4 phosphorylation site(s) regulated by membrane potential. First, we transfected CG neurons with Sp4 containing an N-terminal FLAG tag (FLAG-Sp4), and incubated the cells in 25mM or 5mM KCl as described above (Fig. 2A). We observed a phosphatase sensitive mobility shift in that was induced upon culture in resting conditions, indicating that FLAG-Sp4 is likely to be regulated by the same signaling pathways as endogenous Sp4. To identify specific regions of the Sp4 protein that were required to observe the membrane potential-dependent shift in protein mobility, we generated FLAG-Sp4 N-terminal deletion mutants and analyzed them by immunoblot. The deletion mutants continued to exhibit induced changes in mobility in resting conditions, even when a protein lacking the first 609 N-terminal amino acids and consisting only of amino acids 610–784 was used (Fig. 2B). In contrast, a deletion mutant lacking the C-terminal 54 amino acids did not shift in response to culture in resting conditions (Fig. 2B). Similar to full-length Sp4, the FLAG-Sp4 610–784 deletion mutant shift was sensitive to phosphatase treatment (Fig. 2C). These data suggest that a site of phosphorylation regulated by membrane potential resides within the C-terminal 54 amino acids.
Figure 2. The C-terminal 54 amino acids of Sp4 are required for phosphorylation in resting conditions.
A. Top – Diagram of full-length FLAG-Sp4. Sp4 protein domains are defined as follows: S/T – serine/threonine rich region; Q – glutamine rich region; DBD – DNA binding domain. Bottom – FLAG-Sp4 was transfected into CG neurons and cells were exposed to 25mM (depolarizing) or 5mM (resting) KCl for one hour. Lysates were treated with phosphatase (PPase), separated by 6% PAGE, and analyzed by Western blot. Results are a representative immunoblot. B. Left - Diagram of FLAG-Sp4 deletion mutants transfected into CG neurons and assayed for reduced mobility in resting conditions. Right – A representative immunoblot of each deletion mutant. Lysates were separated by 6% PAGE for 1–784 and 1–730; 8% PAGE for 373–784 and 494–784; and 10% PAGE for 610–784, and the positions of molecular weight markers are indicated. C. Top – Diagram of truncated FLAG-Sp4 610–784. Bottom - Truncated FLAG-Sp4 610–784 was transfected into CG neurons and cells were treated as in (A). Lysates were separated by 10% PAGE and analyzed by Western blot. Results are a representative immunoblot. D. Purified Sp4 from rat cerebellum was analyzed by mass spectrometry, and phospho-peptides were identified corresponding to human S136 and S770.
To identify specific sites of phosphorylation we immunoprecipitated Sp4 from nuclear extracts prepared from P6 rat cerebella, and the purified protein was analyzed by LC/MS/MS for phosphorylated peptides. We detected phosphorylated peptides at sites corresponding to human S136 and S770 (Fig. 2D). Since S770 lies within the region required to observe a shift in Sp4 mobility in resting conditions, we further investigated the regulation and function of Sp4 phosphorylation at S770.
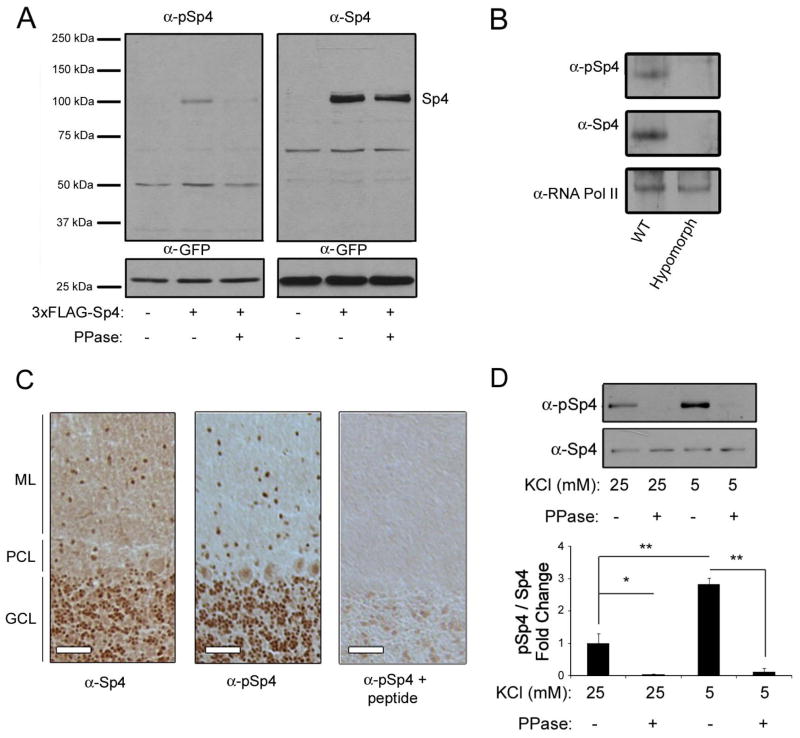
In order to study Sp4 phosphorylation at S770, we generated a phospho-specific antibody. To confirm the antibody bound specifically to Sp4 phosphorylated at S770, we first overexpressed FLAG-Sp4 in 293T cells (Fig. 3A). Both anti-Sp4 and the phospho-Sp4 antibody detected a band at the molecular weight corresponding to FLAG-Sp4. Furthermore, when the lysates were treated with phosphatase, the phospho-Sp4 signal was reduced whereas the total Sp4 signal was unchanged. To ensure the antibody recognized endogenous phospho-Sp4 and did not also recognize a non-specific protein of similar molecular weight, we immunoblotted against cerebellar lysates from wild-type and Sp4 hypomorph mice which express greatly reduced levels of Sp4 (Zhou et al. 2005). Both anti-Sp4 and anti-phospho-Sp4 antisera recognized bands of similar molecular weight in lysates from wild-type mice, but there was no immunoreactivity with either antisera in lysates from hypomorph mice (Fig. 3B). Finally, we examined phospho-Sp4 S770 expression in tissue sections from adult rat cerebellum. We observed largely overlapping expression between Sp4 and phospho-Sp4 S770 (Fig. 3C). Co-incubation of brain slices with the phospho-Sp4 antibody and the phosphorylated peptide antigen eliminated the phospho-Sp4 signal, providing another control for the specificity of the antibody. Together, these results show the phospho-Sp4 antibody exhibits immunoreactivity to exogenous and endogenous Sp4, fails to detect a signal in the Sp4 hypomorph, displays a similar tissue expression pattern as that of a total Sp4 antibody, is competed for binding by a phosphorylated Sp4 peptide, and does not bind to Sp4 in lysates treated with a phosphatase. These results validate the specificity of the phospho-Sp4 antibody in binding to the phosphorylated S770 residue, and support the use of this antibody to study Sp4 S770 phosphorylation.
Figure 3. A phospho-specific antibody identifies Sp4 S770 as a site of phosphorylation regulated by membrane depolarization.
A. 293T cells were transfected with FLAG-Sp4 or an empty vector and lysates were treated with or without lambda phosphatase (PPase) before being separated by 6–12% PAGE and immunoblotted for pSp4 and Sp4. B. Cerebellar lysates from wild-type and Sp4 hypomorph mice were separated by 6% PAGE and immunoblotted with antisera specific for phospho-Sp4 S770 (pSp4), Sp4, and RNA polymerase II as a loading control. C. Cerebellar sections from adult rat brains were subjected to immunohistochemical staining using antibodies to Sp4, pSp4, and pSp4 co-incubated with peptide antigen as indicated. ML – molecular layer; PCL – Purkinje cell layer; GCL – granule cell layer. Scale bar is 50μm. D. CG neurons were exposed to depolarizing or resting conditions for 1 hour and lysates were subjected to phosphatase treatment, separated by 10% PAGE, and analyzed by Western blot using pSp4 and total Sp4 antibodies. Top - representative immunoblot. Bottom - quantification of the ratio of phosphorylated Sp4 relative to total Sp4 and normalized to the control condition. N=3. **p<0.01, *p<0.05, ANOVA (F=47.54; p<0.0001).
CG neuron cultures were then exposed to depolarizing or resting conditions and the level of Sp4 S770 phosphorylation was assessed by Western blot. We found that Sp4 phosphorylation at S770 was strongly induced in resting conditions (Fig. 3D, compare lanes 1 and 3). As an additional control, the phosphorylated Sp4 signal was completely abolished when lysates were treated with a phosphatase (Fig. 3C, lanes 2 and 4). Taken together, our results identify S770 on the transcription factor Sp4 as a site of phosphorylation that is regulated by membrane potential.
Signaling pathways regulating Sp4 phosphorylation at S770
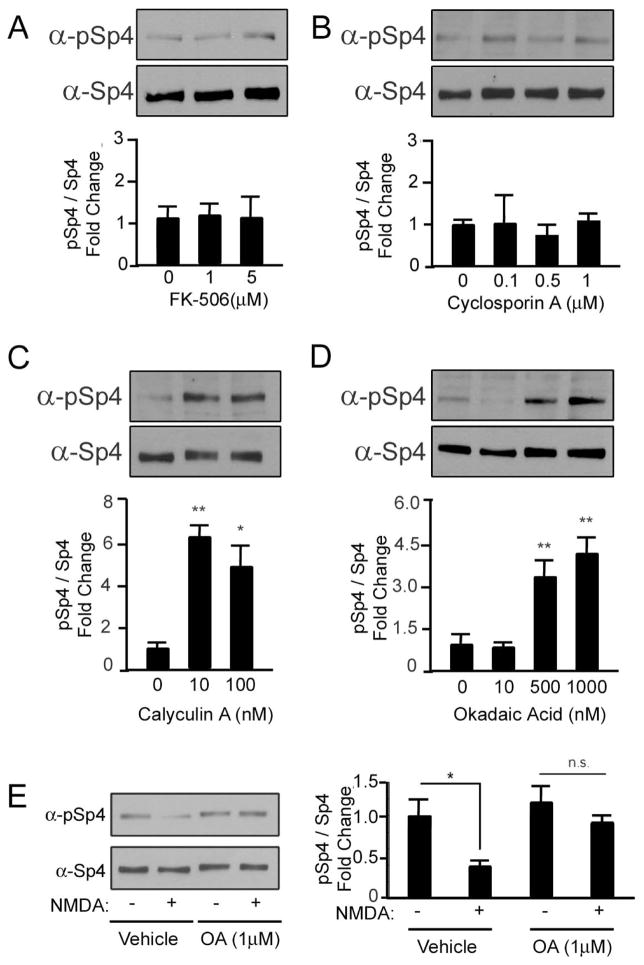
We reasoned that Sp4 S770 phosphorylation was decreased in depolarizing conditions due to signaling pathways activated by a voltage-gated calcium channel. We therefore tested the effect of inhibitors to channels regulated by depolarization and known to signal to transcription factors in the nucleus. In conditions of depolarization, neither the addition of the L-type calcium channel inhibitor, nimodipine, nor the AMPA receptor inhibitor, CNQX, had any effect on Sp4 S770 phosphorylation. In contrast, two different NMDA receptor inhibitors, MK-801 and APV, resulted in a significant increase in Sp4 S770 phosphorylation (Fig. 4A). In contrast to culture in resting conditions, we found that NMDA receptor inhibition had no effect on total Sp4 protein levels (Fig. 4B). These results suggest that Sp4 phosphorylation and degradation are separable processes, a conclusion further supported by our finding that a phospho-mimetic point mutant did not reduce FLAG-Sp4 steady state levels (Fig. S1A). When we placed CG neurons in a magnesium-free solution and treated the cells with 100μM NMDA, which specifically activates NMDA receptors, the levels of Sp4 phosphorylation were also reduced (Fig. 4C). These results indicate that signaling through the NMDA receptor reduces the levels of Sp4 phosphorylation at S770.
Since we observed the expression of phosphorylated Sp4 throughout the brain (data not shown), we tested whether Sp4 phosphorylation is similarly regulated in cultured cortical neurons. We found that depolarization with high KCl or stimulation with 100μM NMDA reduced the levels of Sp4 S770 phosphorylation in cortical neurons (Fig. 4D). These data suggest that NMDA receptor dependent signaling promotes the dephosphorylation of Sp4 S770 in neurons throughout the brain.
Based upon these findings, we hypothesized that the NMDA receptor activates a protein phosphatase to dephosphorylate Sp4. Calcineurin is a phosphatase known to be downstream of the NMDA receptor, and was a logical candidate phosphatase. We failed to see changes in Sp4 phosphorylation, however, when CG neurons were treated with either of two different calcineurin inhibitors: FK-506 or Cyclosporin A (Fig. 5A, B). The PP1 phosphatase is also activated downstream of the NMDA receptor (Morishita et al. 2001). Calyculin A potently inhibits both PP1 and PP2A, and increased the levels of Sp4 phosphorylation (Fig. 5C). Okadaic acid (OA) inhibits PP2A at low concentrations and PP1 at higher concentrations (Sala et al. 2000). When we treated CG neurons with high, but not low, concentrations of OA the level of Sp4 phosphorylation was increased (Fig. 5D). Furthermore, the NMDA-dependent reduction in Sp4 phosphorylation was prevented in the presence of high concentrations of OA (Fig. 5E). These data suggest that activation of the NMDA receptor initiates a PP1/PP2A-dependent signaling pathway to promote the dephosphorylation of Sp4.
Figure 5. The PP1/PP2A phosphatase reduces Sp4 phosphorylation at S770.
CG neurons were treated with the calcineurin phosphatase inhibitors FK-506 (A) or Cyclosporin A (B) or the PP1/PP2A phosphatase inhibitors Calyculin A (C) or Okadaic Acid (D) at the indicated concentrations for one hour and the levels of phospho-Sp4 S770 relative to total Sp4 were analyzed by Western blot and quantified. N=3–5, *p<0.05, **p<0.01; Calyculin A: ANOVA (F=16.20; p=0.0038); Okadaic acid: ANOVA (F= 6.225; p=0.0018). E. CG neurons were stimulated with 100μM NMDA in the presence of vehicle or 1μM Okadaic acid for one hour, and Sp4 phosphorylation at S770 relative to total Sp4 was determined by Western blot. Left - representative immunoblot. Right - quantification. N=3, *p<0.05, n.s., not significant, unpaired T-test (p=0.0183).
Characterization of the function of Sp4 phosphorylation at S770
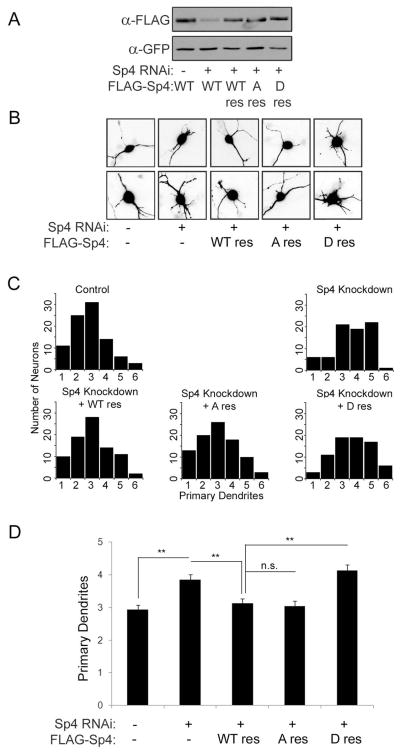
We have shown that Sp4 is phosphorylated, but not degraded, upon NMDA receptor inhibition. This suggests that S770 phosphorylation may function to regulate Sp4 activity independently of protein degradation. Sp4 is required to eliminate excess dendrites during CG neuron development, therefore we investigated if S770 phosphorylation contributes to this function (Ramos et al. 2007). When we overexpressed wild-type and FLAG-Sp4 point mutants, we failed to see changes in CG neuron primary dendrites, consistent with our previous report and possibly due the presence of endogenous Sp4 (data not shown). We therefore generated RNAi resistant FLAG-Sp4 (WT res), as well as serine to alanine non-phosphorylatable (S770A res) and serine to aspartic acid phospho-mimetic (S770D res) point mutants. These mutants were expressed at similar levels in CG neurons, and restored expression in the presence of Sp4 RNAi in heterologous cells (Fig. S1A and Fig. 7A). Furthermore, both the S770A and S770D point mutants functioned similarly to wild-type FLAG-Sp4 to activate a TrkC-luciferase reporter in heterologous cells, suggesting that mutations at this residue did not globally disrupt protein structure or function (Fig. S1B).
CG neurons were cotransfected at DIV 2 with Sp4 RNAi and FLAG-Sp4 rescue constructs together with GFP to visualize transfected neurons and Bcl-xL to eliminate cell viability as confounding factor affecting dendrite morphology (Bcl-xL has little or no effect on dendrite morphology: (Gaudilliere et al. 2004)). At DIV 5, CG neurons transfected with the indicated plasmids were analyzed for the number of primary dendrites, and representative images are shown (Fig. 7B). When we plotted the distribution of dendrite numbers in each group we observed that, as expected, Sp4 knockdown led to a distribution that was skewed towards increased numbers of primary dendrites (Fig. 7C). In these conditions, the number of immature neurons, defined as having greater than 3 dendrites, increased from about 25% to over 50% upon Sp4 knockdown. Both the dendrite distribution and the fraction of immature neurons was restored to control levels by expression of RNAi-resistant WT and S770A Sp4. The phospho-mimetic S770D, however, failed to restore the Sp4-dependent defect in dendrite pruning. When we analyzed the average number of dendrites in each condition, we found that neurons in the control condition had between 2 and 3 primary dendrites, which was significantly increased upon Sp4 knockdown. This knockdown phenotype was rescued when neurons were co-transfected with RNAi-resistant WT and S770A, but not S770D Sp4 (Fig. 7D). These data indicate that the phospho-mimetic mutant impairs the Sp4-dependent maturation of CG neurons, suggesting that phosphorylation of Sp4 at S770 inhibits this function.
Discussion
We present three major findings in this work: First, we identify a novel site of phosphorylation on transcription factor Sp4 at S770. Second, using a phospho-specific antibody we find that Sp4 phosphorylation at S770 is reduced by the activation of the NMDA receptor through a PP1-dependent signaling pathway. Third, we provide evidence that phosphorylation impairs the function of Sp4 in promoting the developmental maturation of CG neuron dendrites.
The role of NMDA receptor signaling in neuronal development is largely context dependent. Activation of the receptor has been shown to promote dendrite outgrowth in some circumstances (Sin et al. 2002, Lei et al. 2006, Sepulveda et al. 2010), and to enhance dendrite elimination in others (Espinosa et al. 2009, Datwani et al. 2002, Lee et al. 2005, Monnerie et al. 2003). The NMDA receptor regulates multiple transcription factors to alter cellular gene expression programs, and it is likely that the specific regulation of these transcription factors contributes to the varied physiological outcomes of receptor activation (Lyons & West 2011). Although degradation of Sp4 has been reported in response to excitotoxic glutamate receptor activation (Mao et al. 2007), we show here for the first time that Sp4 is a transcription factor regulated specifically by NMDA receptor signaling in non-excitotoxic conditions. Since Sp4 activity is also context dependent, our data support the view that the specific downstream signaling from the NMDA receptor may influence the outcome of Sp4 activity.
Sp4 has been implicated in the regulation of dendrite patterning, the induction of long-term potentiation, contributions to behavior including learning and memory, as well as psychiatric disorders such as bipolar disorder and schizophrenia (Ramos et al. 2007, Zhou et al. 2007, Zhou et al. 2005, Zhou et al. 2010, Zhou et al. 2009, Pinacho et al. 2011). The NMDA receptor has also been linked to many of these processes (Paoletti et al. 2013). In fact, altered NMDA receptor signaling was suggested to contribute to some phenotypes in Sp4 mutant mice, as reduced levels of the GluN1 subunit were observed in these animals (Zhou et al. 2010). Based on our data we hypothesize that the modification state of Sp4, informed by NMDA receptor signaling, influences Sp4 transcriptional activity and ultimately contributes to NMDA receptor dependent dendrite elimination.
Our results also implicate the NMDA receptor dependent activation of the PP1/PP2A phosphatase in the regulated dephosphorylation of Sp4. Due to the pharmacological and molecular-genetic limitations of studying phosphatases we cannot rule out a contribution of PP2A to the dephosphorylation of Sp4, however, our data strongly implicate the activity of PP1. PP1 has been shown to specifically contribute to certain forms of NMDA receptor-dependent long-term depression (Genoux et al. 2002, Mulkey et al. 1994, Morishita et al. 2001). NMDA receptor activation of PP1 is often observed in response to stimulation paradigms that activate long-term depression and require calcineurin activation. Our data suggest a calcineurin-independent pathway regulating PP1 activity in CG neurons. Calcineurin-independent activation of PP1 downstream of the NMDA receptor has also been observed, although the specific mechanisms mediating this activation are currently unknown (Sala et al. 2000). An important unanswered question is the identity of the kinase regulating Sp4 S770 phosphorylation. Experiments using pharmacological inhibitors targeting predicted candidate kinases have not yet identified the relevant kinase, and this remains an important ongoing effort.
We show here that both membrane depolarization and NMDA receptor signaling reduce Sp4 S770 phosphorylation. Only in the absence of membrane depolarization, however, is Sp4 protein degradation observed, as inhibition of the NMDA receptor did not reduce Sp4 protein levels (Fig. 4B). These observations suggest that Sp4 phosphorylation at S770 alone is not sufficient to mediate Sp4 protein degradation. In support of this, we did not observe changes in steady-state expression of full-length FLAG-Sp4 when S770 was mutated to alanine or aspartic acid (Fig. S1A). The regulation of Sp4 levels and activity by multiple cell signaling pathways likely allows for more precise control of this transcription factor. Given our finding of an Sp4 phosphorylation site at S136, it is highly likely that additional post-translational modifications contribute to regulating the activity and stability of Sp4.
We investigated the function of Sp4 phosphorylation at S770 using point mutants. We provide evidence that while the non-phosphorylatable S770A mutant was functional, the phospho-mimetic S770D mutant failed to promote CG neuron dendritic maturation. Phospho-mimetics are often useful to assay the function of phosphorylation, however it is a concern that a mutation may destroy the function of the protein or that the mutant will not recapitulate the true activity of a phospho-protein (Tarrant et al. 2012). We observed that both the S770A and S770D mutants were expressed and had a similar ability as wild-type to activate a reporter gene, indicating that many Sp4 functions were preserved in the phospho-mimetic mutant (Fig. S1B). Thus, while the exact mechanism by which Sp4 phosphorylation at S770 impairs CG neuron maturation is unclear, we propose that it involves the failure to activate a specific subset of Sp4 target genes. Since there are currently very few described target genes of Sp4, further studies are needed to identify the specific genes regulating dendrite pruning.
In conclusion, the present study identifies a signal-dependent phosphorylation of the transcription factor Sp4 at S770 that impairs the Sp4-dependent maturation of CG neurons and is regulated by an NMDA-receptor/PP1 signaling pathway. Our results describe a new transcriptional component downstream of NMDA receptor signaling, and also suggest that the phosphorylation state of Sp4 is a mechanism regulating Sp4 activity. These results expand our understanding of the signal-dependent mechanisms regulating neuronal gene expression, which have broad implications for neuronal development and disease.
Supplementary Material
Figure 6. A phospho-mimetic Sp4 S770D fails to rescue Sp4-knockdown dependent deficits in dendrite patterning in cerebellar granule neurons.
A. Extracts from Neuro2A cells cotransfected with control or Sp4 shRNA, along with RNAi-resistant FLAG-Sp4 wild-type (WT res), S770A (A res), or S770D (D res) and GFP were analyzed by Western blot. Results are a representative immunoblot from an experiment performed three times. B. Representative images of CG neurons transfected with the indicated constructs and analyzed by immunofluorescent microscopy for GFP. C. Histograms depicting the number of neurons with the indicated number of dendrites that were observed in each condition. D. Quantification of average primary dendrite numbers from the experiment described in (B). N=80–90, **p<0.01, ANOVA (F=12.01; p<0.0001).
Acknowledgments
This work was supported by: NIH RO1 HD043364 to G.G. and NIH NINDS T32 NS061764 training grant in support of G.S.
Abbreviations used
- AMPA
alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- APV
DL-2-Amino-5-phosphonopentanoic acid
- CG
cerebellar granule
- CNQX
6-cyano-7-nitroquinoxaline-2,3-dione
- FLAG-Sp4
N-terminal FLAG-tagged Sp4
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- LC/MS/MS
Liquid chromatography tandem mass spectrometry
- OA
okadaic acid
- PAGE
Polyacrylamide gel electrophoresis
- PP1/PP2A
protein phosphatase 1/2A
- pSp4
Sp4 phosphorylated at S770
- S770A res
RNAi resistant FLAG-Sp4 with a serine to alanine point mutation at amino acid 770
- S770D res
RNAi resistant FLAG-Sp4 with a serine to alanine point mutation at amino acid 770
- SDS–PAGE
sodium dodecyl sulfonate–polyacrylamide gel electrophoresis
- WT res
RNAi resistant FLAG-Sp4
Footnotes
Conflict of interest disclosure
The authors do not declare a conflict of interest.
References
- Bilimoria PM, Bonni A. Cultures of cerebellar granule neurons. CSH protocols. 2008;2008 doi: 10.1101/pdb.prot5107. pdb prot5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Uren JM, Kittler JT, Wang H, Olsen R, Parker PJ, Moss SJ. Subunit-specific association of protein kinase C and the receptor for activated C kinase with GABA type A receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19:9228–9234. doi: 10.1523/JNEUROSCI.19-21-09228.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Greenberg ME. Communication between the synapse and the nucleus in neuronal development, plasticity, and disease. Annual review of cell and developmental biology. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwani A, Iwasato T, Itohara S, Erzurumlu RS. NMDA receptor-dependent pattern transfer from afferents to postsynaptic cells and dendritic differentiation in the barrel cortex. Molecular and cellular neurosciences. 2002;21:477–492. doi: 10.1006/mcne.2002.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, Wheeler DG, Tsien RW, Luo L. Uncoupling dendrite growth and patterning: single-cell knockout analysis of NMDA receptor 2B. Neuron. 2009;62:205–217. doi: 10.1016/j.neuron.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudilliere B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418:970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Hardingham GE. Coupling of the NMDA receptor to neuroprotective and neurodestructive events. Biochemical Society transactions. 2009;37:1147–1160. doi: 10.1042/BST0371147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Ramu S, Lee S, Aguilar B, Ganesan SK, Yoo J, Kalra VK, Koh CJ, Hong YK. Phosphate-buffered saline-based nucleofection of primary endothelial cells. Analytical biochemistry. 2009;386:251–255. doi: 10.1016/j.ab.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Molecular cell. 2002;9:1005–1016. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- Lee LJ, Lo FS, Erzurumlu RS. NMDA receptor-dependent regulation of axonal and dendritic branching. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:2304–2311. doi: 10.1523/JNEUROSCI.4902-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z, Ruan Y, Yang AN, Xu ZC. NMDA receptor mediated dendritic plasticity in cortical cultures after oxygen-glucose deprivation. Neuroscience letters. 2006;407:224–229. doi: 10.1016/j.neulet.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Progress in neurobiology. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Yang SH, Simpkins JW, Barger SW. Glutamate receptor activation evokes calpain-mediated degradation of Sp3 and Sp4, the prominent Sp-family transcription factors in neurons. Journal of neurochemistry. 2007;100:1300–1314. doi: 10.1111/j.1471-4159.2006.04297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnerie H, Shashidhara S, Le Roux PD. Effect of excess extracellular glutamate on dendrite growth from cerebral cortical neurons at 3 days in vitro: Involvement of NMDA receptors. Journal of neuroscience research. 2003;74:688–700. doi: 10.1002/jnr.10797. [DOI] [PubMed] [Google Scholar]
- Morishita W, Connor JH, Xia H, Quinlan EM, Shenolikar S, Malenka RC. Regulation of synaptic strength by protein phosphatase 1. Neuron. 2001;32:1133–1148. doi: 10.1016/s0896-6273(01)00554-2. [DOI] [PubMed] [Google Scholar]
- Mulkey RM, Endo S, Shenolikar S, Malenka RC. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature reviews Neuroscience. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Pinacho R, Villalmanzo N, Lalonde J, Haro JM, Meana JJ, Gill G, Ramos B. The transcription factor SP4 is reduced in postmortem cerebellum of bipolar disorder subjects: control by depolarization and lithium. Bipolar disorders. 2011;13:474–485. doi: 10.1111/j.1399-5618.2011.00941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinacho R, Villalmanzo N, Roca M, et al. Analysis of Sp transcription factors in the postmortem brain of chronic schizophrenia: A pilot study of relationship to negative symptoms. Journal of psychiatric research. 2013;47:926–934. doi: 10.1016/j.jpsychires.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos B, Valin A, Sun X, Gill G. Sp4-dependent repression of neurotrophin-3 limits dendritic branching. Molecular and cellular neurosciences. 2009;42:152–159. doi: 10.1016/j.mcn.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Rudolph-Correia S, Sheng M. Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suzuki K, Nakanishi S. NMDA receptor stimulation and brain-derived neurotrophic factor upregulate homer 1a mRNA via the mitogen-activated protein kinase cascade in cultured cerebellar granule cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:3797–3805. doi: 10.1523/JNEUROSCI.21-11-03797.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulveda FJ, Bustos FJ, Inostroza E, Zuniga FA, Neve RL, Montecino M, van Zundert B. Differential roles of NMDA Receptor Subtypes NR2A and NR2B in dendritic branch development and requirement of RasGRF1. Journal of neurophysiology. 2010;103:1758–1770. doi: 10.1152/jn.00823.2009. [DOI] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Molecular psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyn SI, Shi J, Kraft JB, et al. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Molecular psychiatry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Tam GW, van de Lagemaat LN, Redon R, et al. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochemical Society transactions. 2010;38:445–451. doi: 10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- Tarrant MK, Rho HS, Xie Z, et al. Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nature chemical biology. 2012;8:262–269. doi: 10.1038/nchembio.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Molecular psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- Zhou X, Nie Z, Roberts A, Zhang D, Sebat J, Malhotra D, Kelsoe JR, Geyer MA. Reduced NMDAR1 expression in the Sp4 hypomorphic mouse may contribute to endophenotypes of human psychiatric disorders. Human molecular genetics. 2010;19:3797–3805. doi: 10.1093/hmg/ddq298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Qyang Y, Kelsoe JR, Masliah E, Geyer MA. Impaired postnatal development of hippocampal dentate gyrus in Sp4 null mutant mice. Genes, brain, and behavior. 2007;6:269–276. doi: 10.1111/j.1601-183X.2006.00256.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Tang W, Greenwood TA, Guo S, He L, Geyer MA, Kelsoe JR. Transcription factor SP4 is a susceptibility gene for bipolar disorder. PloS one. 2009;4:e5196. doi: 10.1371/journal.pone.0005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.