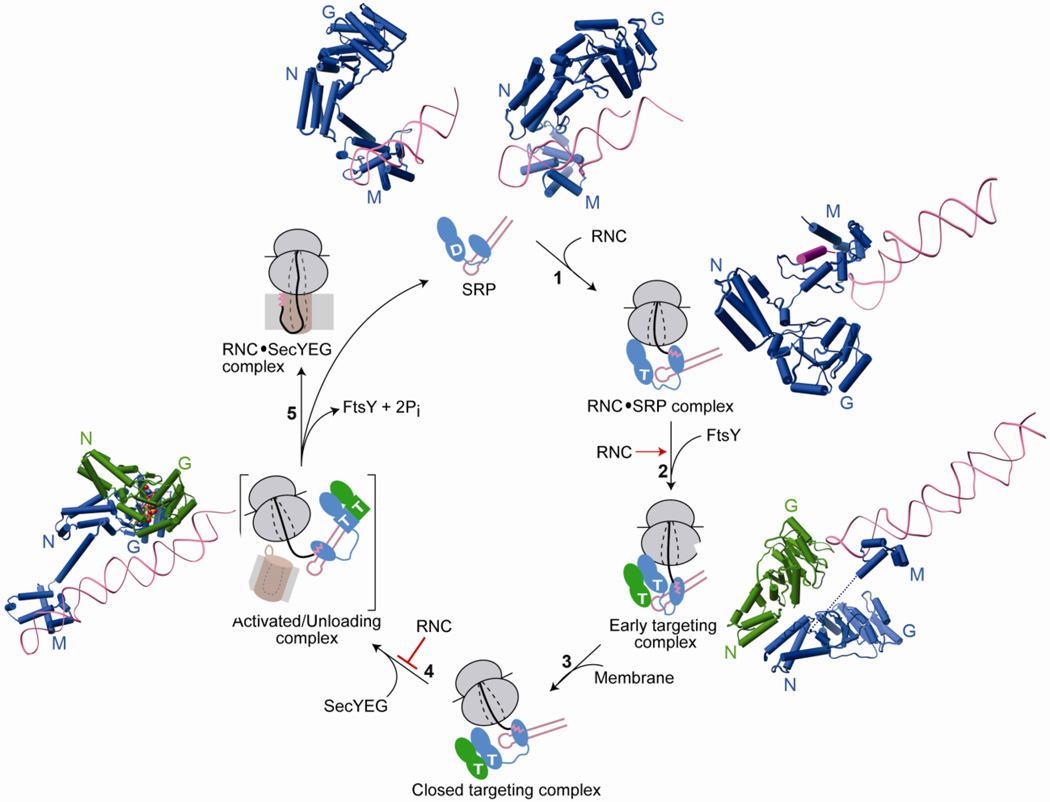
Fig. 4.
Conformational changes in SRP and SR GTPases are coupled to global reorganization of the SRP particle and are regulated by biological effectors for the pathway. Free SRP exists in a number of conformations in which the NG-domain of Ffh is oriented differently with respect to the M-domain and the SRP RNA. The top panel shows structures of SRP from S. solfataricus (PDB ID: 1qzw, left) and M. jannaschii (PDB ID: 2v3c, right) highlighting its conformational flexibility. The binding of RNC to SRP favors an SRP conformation in which the tetraloop of the SRP RNA is poised to interact with the G-domain of SR (step 1). This interaction strongly stabilizes the early targeting complex resulting in very efficient assembly of this complex (step 2). Top right (PDB ID: 2j28) and bottom right (PDB ID: 2xkv) panels show molecular models of the interaction of RNC with SRP without or with FtsY. Anionic phospholipids in the membrane strongly accelerate the rearrangement of the early targeting complex to the closed state (step 3). Interaction with SecYEG induces the SRP/SR complex into the activated state (step 4) in which the NG-domain complex relocalizes to the distal end of SRP RNA (left panel, PDB ID: 2xxa). This movement is negatively regulated by the RNC allowing a productive search for the translocon. The activated complex is shown in brackets to indicate that it is a proposed intermediate with transient lifetime, and its precise structure is not known. Hydrolysis of GTP triggers disassembly of the GTPase complex while the cargo is transferred to the translocon (step 5).