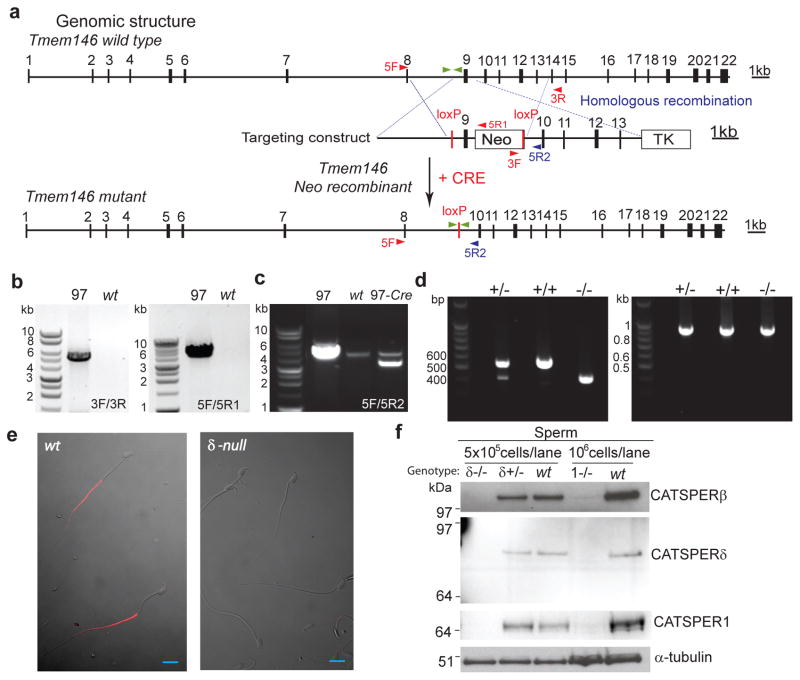
Figure 4. Targeted disruption of Tmem146 gene inactivates CatSperδ.
(a) Whole genomic structure of mouse Tmem146 gene, targeting construct, and predicted mutant allele generated by homologous recombination and Cre excision; Filled boxes, exons; thin lines, introns. Neomycin (Neo) and thymidine kinase (TK) selection cassettes are shown. The locations of primers used for genotyping are shown with arrowheads. Primers located outside targeting region, 5F and 3R; inside targeting region, 5R2; within Neo, 5R1 and 3F. (b and c) PCR genotyping of genomic DNA. Tmem146 Neo mutant ES clone (97) is identified by the presence of PCR products at both the 3′- (3F/3R) and 5′- (5F/5R2) of the expected locus (b). Excision of genomic sequence flanked by two loxP sites by PCR with 5F/5R2 (c). (d) RT-PCR of CatSperδ with testis mRNA from CatSperδ–null mice showing the expected products. Combination of two primers with one spanning exon 7/8 and the other in exon 12 demonstrates absence of exon 9 in the CatSperδ transcript (left). RT-PCR of GAPDH is the control (right), CatSperδ null–(−/−), CatSperδ–heterozygotes (+/−) and wildtype (+/+). (e) Immunostaining of mouse epididymal sperm with anti-CATSPERδ antibody (α-δ446). CATSPERδ specifically labels only the principal piece of the tail of wt spermatozoa (left) and is not observed in the CatSperδ-null sperm cells (right). Scale bar, 10 μm. (f) Absence of CATSPER1, β and δ proteins in spermatozoa of CatSperδ homozygous null mice.