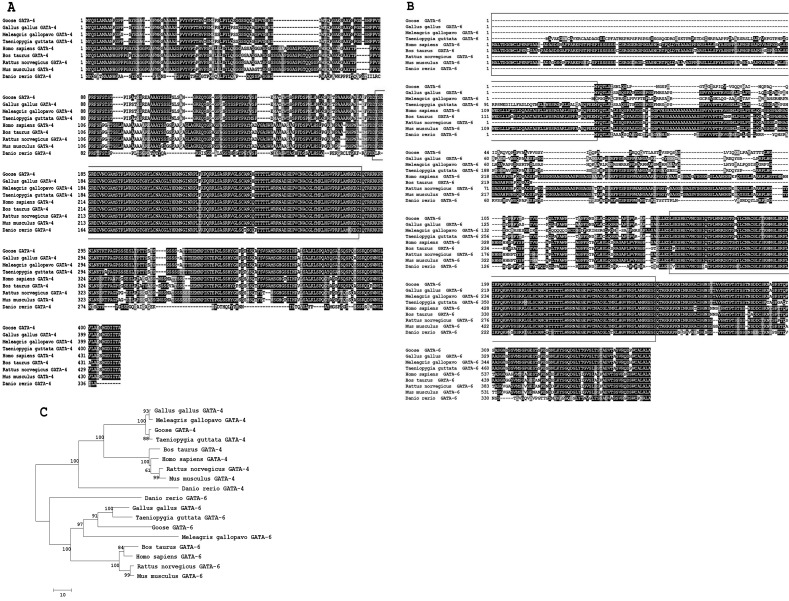
Fig. 1.
Amino acid sequence alignments of GATA-4 and GATA-6 sequences from the goose and other species. The GATA-4 and GATA-6 amino acid sequences used for analysis were extracted from the GenBank database for the following species: Gallus gallus (GATA-4, XP_420041.1; GATA-6, NP_990751.1); Meleagris gallopavo (GATA-4, XP_003204668.1; GATA-6, XP_003205043.1); Taeniopygia guttata (GATA-4, XP_002186842.1; GATA-6, XP_002194999.1); Bos taurus (GATA-4, NP_001179806.1; GATA-6, XP_001253597.2); Homo sapiens (GATA-4, NP_002043.2; GATA-6, NP_005248.2); Rattus norvegicus (GATA-4, NP_653331.1; GATA-6, NP_062058.1); Mus musculus (GATA-4, NP_032118.2; GATA-6, NP_034388.2); and Danio rerio (GATA-4, NP_571311.1; GATA-6, NP_571632.1). The black blocks represent identical residues between the sequences shown, while the red boxes represent zinc finger domains. A: A multiple amino acid sequence alignment of GATA-4. B: A multiple amino acid sequence alignment of GATA-6. The black boxes represent the 5’-upstream sequences in Taeniopygia guttata, Homo sapiens, Bos taurus and Mus musculus, which have an extra amino-terminal extension from the initiator methionine, thus belonging to the long type of GATA-6, and all other species belong to the short type of GATA-6. C: A phylogenetic tree of the amino acid sequences of GATA-4 and GATA-6 was constructed using the neighbor-joining method (1000 bootstrap replicates).