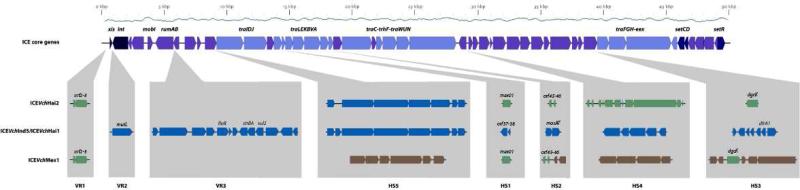
Fig. 1. Structural comparison between ICEVchInd5 (ICEVchHai1), ICEVchHai2 and ICEVchMex1.
The upper map represents the set of core genes common to all SXT/R391 ICEs. Highlighted are genes involved in site-specific excision and integration (xis, int, mobI), error prone DNA repair (rumAB), entry exclusion (eex), ICE regulation (setCDR) and conjugative transfer (tra genes). The bottom map represents the specific regions of ICEVchInd5 (ICEVchHai1), ICEVchHai2, and ICEVchMex1 inserted the variable regions and hotspots. For a detailed description of hotspot 4 in ICEVchHai2 see Table 1.