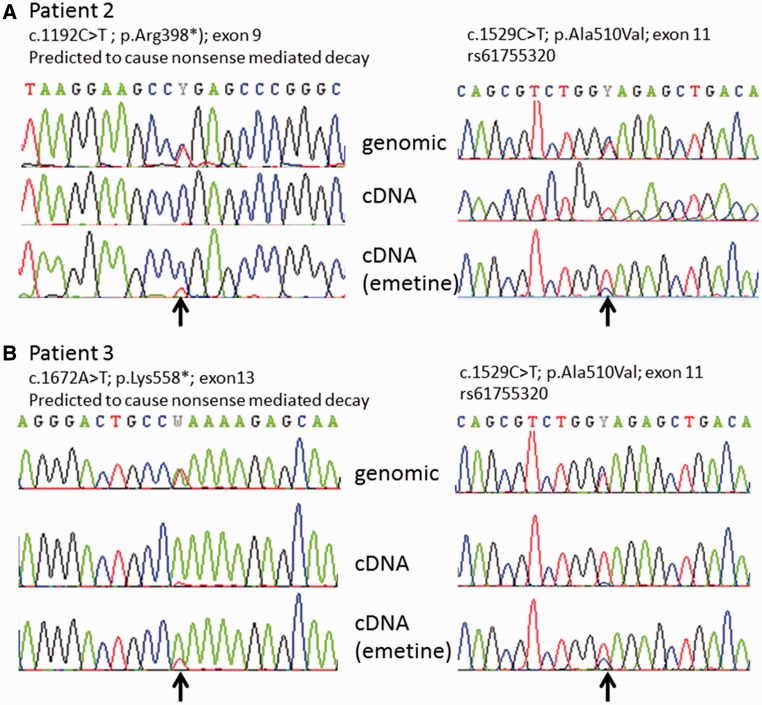
Figure 4.
Confirmatory complementary DNA sequencing in Patients 2 and 3. Arrows indicate the positions of the mutations. (A) In Patient 2, the c.1192C>T (p.Arg398*) mutation is predicted to cause nonsense mediated messenger RNA decay. At left the genomic sequence demonstrates this mutation to be present in the heterozygous state although in complementary DNA from fibroblasts it appears absent because it is degraded by nonsense mediated decay. The presence of this variant is partially restored in fibroblasts grown with emetine (an inhibitor of nonsense mediated decay). The second mutation in this patient [c.1529C>T; p.(Ala510Val)] is present at homozygous levels in complementary DNA indicating that it is on the opposite allele; as before the second allele is partially restored with emetine treatment. (B) Patient 3 has a c.1672A>T (p.Lys558*) mutation which similarly is predicted to cause nonsense mediated decay. The mutation is almost absent in complementary DNA but partially restored in cells grown with emetine. The second c.1529C>T; p.(Ala510Val) mutation is again shown to be present on the opposite allele.