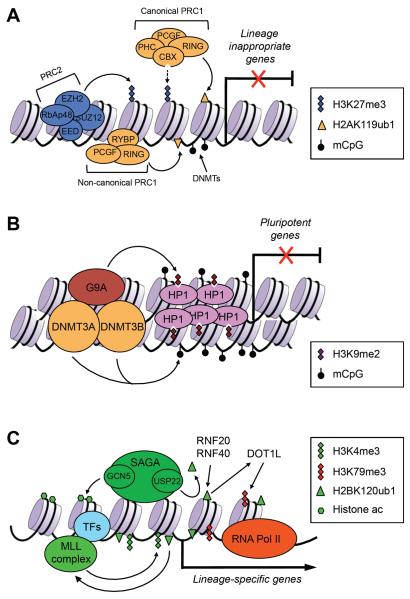
Figure 3. Chromatin regulators involved in gene regulation during postimplantation development and cellular differentiation.
(A) PcG proteins play important roles in repressing developmental genes. The PRC2 complex deposits the repressive H3K27me3 marks, which create binding sites for the canonical, CBX-containing PRC1 complex. The noncanonical, RYBP-containing PRC1 complex binds to chromation via H3K27me3-independent mechanisms. RING1A and RING1B, components of the PRC1 complexes, mediate the repressive H2AK119ub1 modification. DNA methylation, mediated by DNA methyltransferases (DNMTs), is also important in repressing lineage-commitment genes, especially those with low CpG-content promoters. (B) G9A is critical for silencing pluripotency-associated genes in postimplantation embryos and differentiated cells. G9A mediates H3K9me2, which induces heterochromatinization by recruiting HP1. G9A also recruits DNMT3A and DNMT3B, which initiate de novo DNA methylation. (C) In differentiating cells, lineage-specific transcription factors (TFs) recruit chromatin-modifying complexes, such as the MLL complex and the SAGA complex, to lineage-specific genes to create `open' chromatin states. The MLL complex deposits the active H3K4me3 marks. The SAGA complex has at least two enzymatic activities: histone acetylation by GCN5 and H2BK120 deubiquitylation by USP22. H2BK120ub1, mediated by RNF20 and RNF40, is preferentially enriched in the coding regions of differentiation-related genes, but not pluripotency-associated genes. H2BK120ub1 has been shown to promote MLL-mediated H3K4 methylation and DOT1L-mediated H3K79 methylation.