Abstract
Analysis of the individual steps in metastasis is crucial if insights at the molecular level are to be linked to the cell biology of cancer. A technical hurdle to achieving the analysis of the individual steps of metastasis is the fact that, at the gross level, tumors are heterogeneous in both animal models and patients. Human primary tumors show extensive variation in all properties ranging from growth and morphology of the tumor through tumor-cell density in the blood and formation and growth of metastases. Methods capable of the direct visualization and analysis of tumor-cell behavior at single-cell resolution in vivo have become crucial in advancing the understanding of mechanisms of metastasis, the definition of microenvironment, and the markers related to both. This article discusses the use of high-resolution multiphoton imaging of tumors (specifically breast tumors in mice) in vivo.
BACKGROUND
Our primary interest has been in breast tumors because metastasis is dependent on cell motility involving macrophage-mediated chemotaxis (Condeelis and Pollard 2006). Invasive migration and entry of tumor cells into the circulation, common features of breast tumors, are crucial early steps in the metastatic cascade and have been assayed in various ways but only indirectly using conventional methods (Liotta et al. 1974; Butler and Gullino 1975; Glaves 1986).
Human breast carcinomas, in particular, the vast majority of which are ductal carcinomas, are extremely heterogeneous both clinically and pathologically. Current methods of histologic grading, such as the modified Bloom–Richardson method, allow pathologists to separate tumors into three strata. Distinct differences in survival rates, however, effectively apply only to those patients with very well-differentiated or low-grade tumors. Recurrence-free survival for grade 1 carcinomas at 5 and 10 yr, for example, are 75% and 64%, respectively, while the same figures for grade 2 carcinomas are 50% and 40%, and for grade 3 carcinomas are 40% and 38%. This suggests considerable overlap in features of grades 2 and 3 tumors, with classification by grade being a very imperfect means for effective tailoring of management and prediction of outcome (Elston and Ellis 1991; Carter 2001). Gene-expression profiling with microarrays has shown promise in refining these groups further (Ramaswamy et al. 2003) and supports the notion that the invasive and metastatic potentials of the primary tumor are encoded early in the bulk of the tumor, including the stroma (Bernards and Weinberg 2002). However, none of these approaches reveals the mechanistic basis for the differences in the outcome of human breast tumors. Additional approaches that interrogate tumors as to the microenvironments that support invasion and metastasis using common markers in otherwise heterogeneous tumors may reveal mechanisms behind the outcome in sufficient detail to generate new strategies for marker development that can be used in routine histopathological diagnosis, prognosis, and treatment.
MULTIPHOTON INTRAVITAL IMAGING AT SINGLE-CELL RESOLUTION
Within the past several years, imaging methods for the detailed characterization of the behavior of carcinoma cells and macrophages within intact primary tumors using multiphoton imaging combined with fluorescent animal models has become routine (Condeelis and Segall 2003; Wyckoff et al. 2004, 2006, 2007; Sahai et al. 2005; Sidani et al. 2006; Soon et al. 2007; Wang et al. 2007). These methods give information directly about cell behavior in vivo at single-cell resolution and in different microenvironments of the tumor, an essential capability given the heterogeneity of tumors. Imaging is inherently quantitative, allowing the quantification of behavior parameters, including directional migration toward histological landmarks such as blood vessels, frequency, velocity, and persistence of cell motility in vivo, as well as interactions between tumor cells and stromal cells, leading to invasion, intravasation, and extravasation. These imaging methods are valuable in defining (1) cell behaviors that are necessary for invasion, intravasation, and extravasation; (2) cell behavior phenotypes of cells with specific mutations; (3) polarized motility and chemotaxis of cells in vivo; and (4) the definition, size, and regulation of microenvironments in vivo. For more information about multiphoton microscopy and its applications to the study of cell behavior in vivo, see our websites www.einstein.yu.edu/aif/page.aspx and www.einstein.yu.edu/aif/intravital_imaging/introduction.htm.
ADVANTAGES OF MULTIPHOTON MICROSCOPY OVER CONFOCAL MICROSCOPY
We have shown that multiphoton excitation fluorescence microscopy has important advantages over other imaging techniques such as confocal microscopy, particularly for the study of live cells and/or for thick tissues (Farina et al. 1998; Wang et al. 2002). The advantages of the multiphoton instruments include the following.
The lack of fluorophore excitation in regions away from the focal plane minimizes fluorophore bleaching and the generation of toxic by-products during imaging.
Multiphoton images are less prone to degradation by light scattering. This is because the longer wavelengths used for excitation suffer less scattering from microscopic refractive-index differences within the sample, allowing much greater penetration of the tissues (Centonze and White 1998). In addition, as all the resolution is defined by the geometry of the excitation beam, the fluorescence emission is unaffected by light scattering. The reduced sensitivity of multi-photon imaging to light scattering is particularly advantageous for the study of living specimens because of the presence of many refractive-index interfaces in living tissues.
The longer wavelengths used in multiphoton microscopy have the added benefit of second-harmonic generation to image collagen fibers (Campagnola et al. 2001; Williams et al. 2001). This phenomenon is not found in conventional microscopy and allows for imaging of cell-to-matrix interactions such as adhesion and degradation.
A valuable benefit resulting from the second-harmonic-generated image is the ability to directly estimate the amount and condition of the extracellular matrix (ECM) adjacent to carcinoma cells in tumors. The amount, integrity, and size of ECM fibers can be determined by measuring the signal intensity and morphology of fibers in images obtained from the second-harmonic-generated light emitted from the ECM (Wang et al. 2002). An important insight resulting from this analysis is the demonstration that, adjacent to carcinoma cells in the primary tumor, metastatic MTLn3-derived tumors contain two- to threefold less ECM fibers than nonmetastatic MTC-derived tumors (Wang et al. 2002). This is determined by calculating the pixel intensity of a volume using the National Institutes of Health (NIH) Image software. Furthermore, the morphology of the fibers shows discontinuities in second-harmonic emissions along the fibers, suggesting that proteolysis has occurred at intervals along the ECM fibers (Wang et al. 2002; Sidani 2006). Finally, these fibers are observed to support the rapid and directional linear migration of carcinoma cells in metastatic tumors (Wang et al. 2002). This is an exciting observation because chemotaxis of carcinoma cells in response to epidermal growth factor-like (EGF-like) ligands resulting from proteolysis of the ECM could be an important property of invasive tumors (Liotta and Kohn 2001).
The second-harmonic signal is emitted as polarized light when the electrons in the π orbitals of α-helix chains, such as those found in collagens, are excited by a long wavelength of light (Campagnola et al. 2001; Williams et al. 2001). We have found that we are able to excite the second-harmonic signal with wavelengths of 760–960 nm and image them through a filter with a 450–480-nm cutoff.
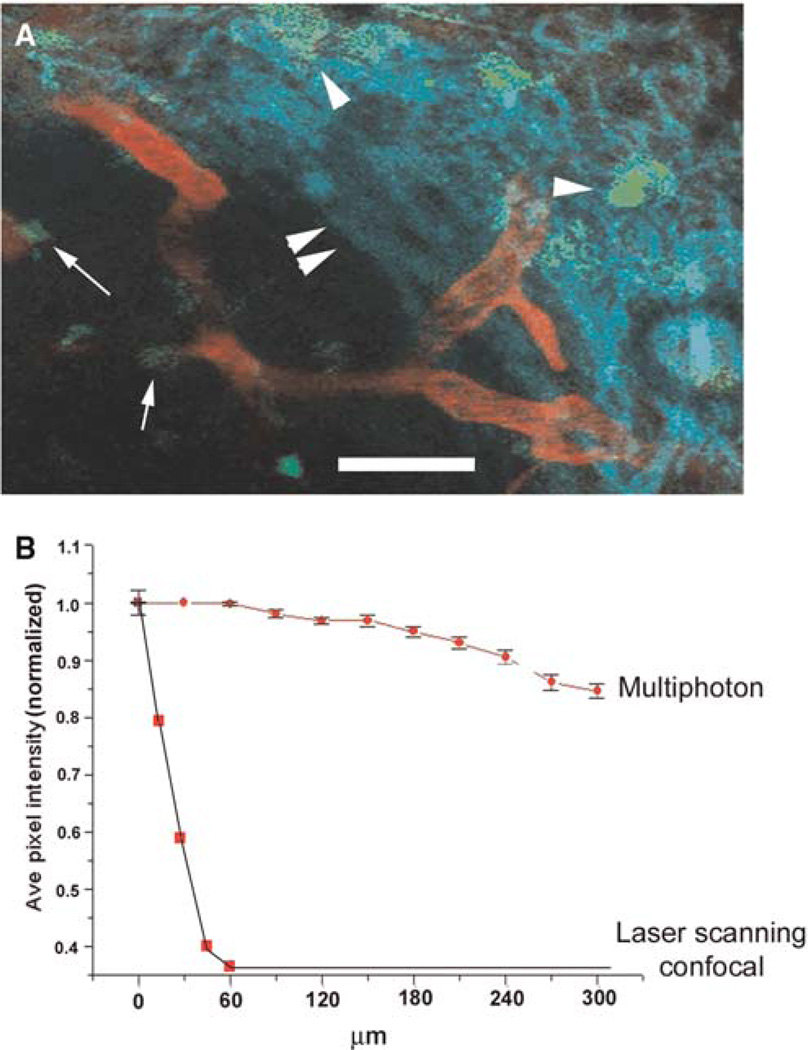
Direct comparisons of confocal imaging with multiphoton imaging have demonstrated that, although the deepest useful image possible with the conventional confocal was <60 μm, useful images were obtained with the multiphoton microscope to depths >300 μm (Wang et al. 2002; Kedrin et al. 2008). In addition, the bleach rate is much less in the multiphoton microscope, and signal to noise does not fall off significantly throughout the z series obtained with the multiphoton (Fig. 1). In both imaging techniques (i.e., in the live animal under anesthesia, on either a laser-scanning confocal microscope or amultiphotonmicroscope), we found that green fluorescent protein (GFP) in the tumor cells is an excellent cytoplasmic volume marker that allows the entire cell outline to be defined in vivo in the living intact tissue without rejection of the tumor due to GFP expression (Farina et al. 1998; Condeelis et al. 2000).
FIGURE 1.
Multiphoton microscopy allows for much deeper penetration into tissue as compared with conventional confocal microscopy. (A) A mouse mammary tumor from an MMTV-PyMT × Cfms-GFP transgenic mouse was imaged using a multiphoton microscope. Macrophages, both stromal (arrowheads) and perivascular (arrows), are seen in green. Collagen (double arrowheads) seen as second-harmonic-generated polarized light is seen in blue. Blood vessels, labeled with Texas red dextran, are seen in red. Scale bar, 75 μm. (B) Multiphoton microscopy can image to 300 μm with very little falloff in signal intensity while almost all signal is gone by 70 μm in conventional laser-scanning confocal microscopy.
ANIMAL MODELS FOR MULTIPHOTON IMAGING: TRANSGENIC MICE
To relate the expression of oncogenes to the progression and metastasis of primary breast tumors, the transgenic mouse has served as an important research tool to assess the tissue-specific action of oncogenes in vivo. Of particular importance are transgenic mouse models of breast cancer that resemble the human disease in both etiology and histology. These mouse models are derived from the expression of either the polyomavirus middle T antigen (PyMT) or neu/erbB-2 under the control of the mammary epithelium-specific mouse mammary tumor virus MMTV promoter. The MMTV promoter has been used in a number of studies to generate expression of proteins in the mammary gland (Gunzburg et al. 1991; Webster and Muller 1994; Böttinger et al. 1997). Expression studies using this promoter to drive β-galactosidase expression indicate that, in female mice, the major site of expression is in the epithelial cells of the mammary gland.
Transgenicmice expressing either normal or activated forms of neu have been generated by Muller and colleagues (Muller et al. 1998; Siegel et al. 1999). Mice harboring the activated transgene (NDL1) develop mammary adenocarcinomas with an average onset of 135 d (±37), and ~57% of those animals develop lung metastases within an average of 45 d after the appearance of the primary tumor. ErbB-2 family members directly interact with a diverse set of signaling proteins such as Src PTKs, Shc, PLC-γ, and PI3K. The PyMT is a related oncogene that exerts its oncogenic effects through its association with Src PTKs. The signaling path from Src onward in PyMT mice is believed to be very similar to that in activated-neu mice (Muller et al. 1998). These models are particularly relevant to the study of human breast cancer because erbB-2 is overexpressed in a large percentage of primary breast cancers and the degree of its overexpression predicts a poor clinical prognosis in both lymph-node-positive and lymph-node-negative patients (Price et al. 1997).
During the past 6 yr, we have developed transgenic mice that express fluorescent protein from tissue-specific promoters crossed with mice that express oncogenes from the MMTV promoter, generating mammary tumors with different fluorescent cell types and thereby making these cells in the primary tumors of the mammary gland visible for imaging at cellular resolution (Ahmed et al. 2002; Condeelis and Segall 2003; Wang et al. 2003; Goswami et al. 2004; Wyckoff et al. 2004a,b, 2007).
The transgenic mouse models created are:
-
1.
MMTV-PyMT × WAP-Cre/CAG-CAT-EGFP or MMTV-Cre/CAG-CAT-EGFP mice
-
2.
MMTV-HER2/neu ×WAP-Cre/CAG-CAT-EGFP or MMTV-Cre/CAG-CAT-EGFP mice
These produced mice with GFP-fluorescent carcinoma cells in PyMT- and HER2/neu-oncogene-generated tumors. These mammary tumors were used to image migratory cells and their accompanying stromal cells. Stromal cells were imaged as shadows because they scattered light from the fluorescent cells in the tumor (Condeelis et al. 2000; Wyckoff et al. 2000; Condeelis and Segall 2003).
-
3.
MMTV-PyMT × lys-GFPKi
-
4.
MMTV-HER2/neu × lys-GFPKi
These produced mice with GFP-fluorescent macrophages in the same types of tumors. These mice were used to confirm that shadow cells are potentially macrophages. Because the lys-GFP mice also express GFP in neutrophils, we also used mice expressing GFP from the CSF-1-receptor promoter, which is more macrophage specific (Sasmono et al. 2003). Similar results were obtained with the CSF-1-receptor promoter-GFP mice.
-
5.
MMTV-PyMT × MMTV-iCre/CAG-CAC-ECFP × c-fms-GFP
-
6.
MMTV-HER2/neu × MMTV-iCre/CAG-CAC-ECFP × c-fms-GFP
To visualize tumor cells and macrophages simultaneously in the same animal, we prepared the two mouse models described above (5 and 6) by breeding mice with genetic susceptibility to mammary tumors expressing cyan fluorescent protein (CFP) (models 1 and 2) with mice expressing GFP in their myelomonocytic cells (from the c-fms promoter):
-
7.
Tie2-GFP × MMTV-PyMT
We obtained Tie2/GFP transgenic mice from Dr. Richard Lang, (Skirball Institute, New York University). Tie2 is expressed specifically in endothelial cells, and consequently, GFP can mark the vasculature at sites of Tie2 expression (Yuan et al. 2000). Because the GFP expression in these mice can be mosaic, we also mark blood vessels using intravenous Texas red dextran (10 kDa to 2 mDa). Similar conclusions were reached in our studies using both methods (Wyckoff et al. 2007).
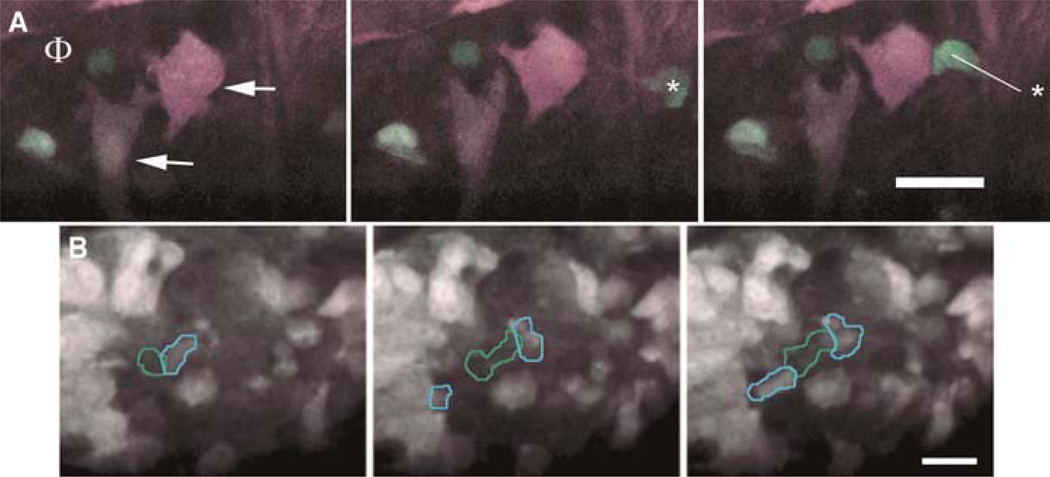
All of these transgenicmouse models with mammary tumors are suitable for high-resolution multiphoton intravital imaging. We have used several together with multiphoton imaging of undissected living primary mammary tumors (reviewed in Condeelis and Segall 2003; Condeelis et al. 2005; Wang et al. 2005; Sidani et al. 2006; Soon et al. 2007). This effort revealed the microenvironments in which tumor cells undergo migration and intravasation in mammary tumors and the importance of macrophages in these events (reviewed in Condeelis and Segall 2003; Condeelis and Pollard 2006; Yamaguchi et al. 2006; Kedrin et al. 2007). In particular, it was found that chemotaxis of tumor cells toward macrophages and their relay chemotaxis to generate single files were essential for invasion in primary mammary tumors of mice (Fig. 2) (Goswami et al. 2005). In addition, it was found that chemotaxis of tumor cells toward perivascular macrophages was essential for intravasation (Fig. 3) (Wyckoff et al. 2007). The point at which tumor cells migrate through the endothelium of blood vessels was identified to be the site of docking on the blood vessel of at least one perivascular macrophage (Fig. 3) (Wyckoff et al. 2007).
FIGURE 2.
CFP tumor cells and GFP macrophages are imaged as they interact in vivo using multiphoton microscopy. (A) In three panels taken from a movie of a tumor from an MMTV-PyMT × MMTV-iCre/CAG-CAC-ECFP × c-fms-GFP transgenic mouse, a perivascular macrophage (Φ) is attracting two tumor cells (arrows) that, in turn, attract another macrophage (* in the third frame) all converging on the perivascular macrophage (Φ). The line shows the extent of macrophage movement between frames. Time between frames is 20 min with each panel being a consecutive frame. (B) In another field, tumor cells (blue outline) and a macrophage (green outline) are seen moving together using signaling from a paracrine loop to form a single file of cells. Scale bars, 25 μm.
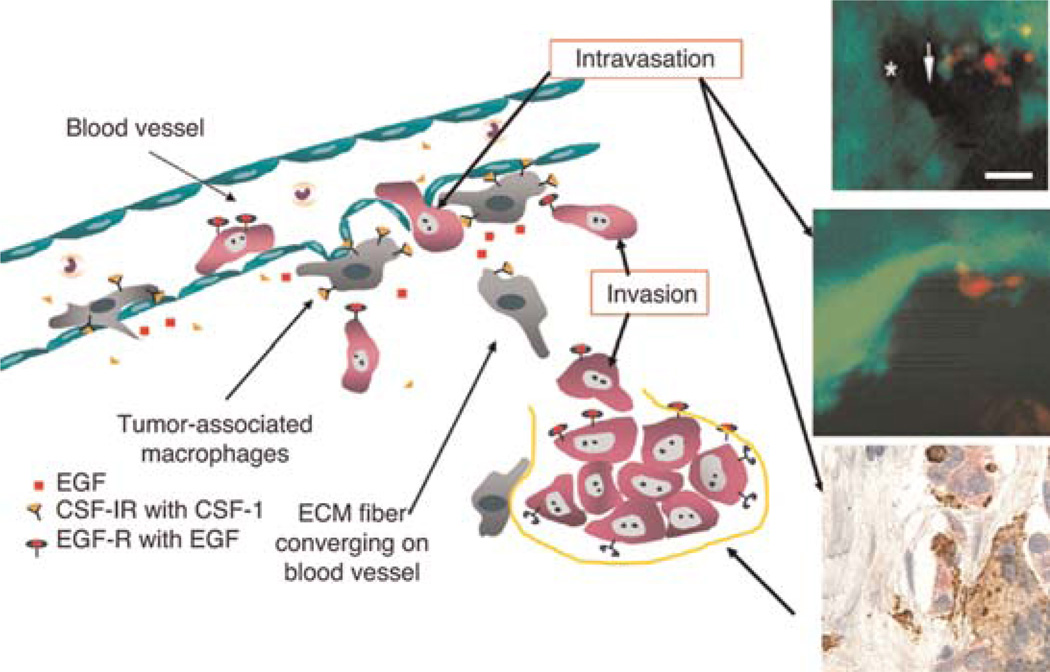
FIGURE 3.
The invasion and intravasation microenvironments discovered using multiphoton microscopy. The cartoon summarizes results from multiphoton imaging defining the invasion and intravasation microenvironments. The top two insets show the interactions of tumor cells (green) and macrophages (red, after Texas-red-dextran phagocytosis) in the intravasation microenvironment in mammary tumors in vivo in mice. The same intravasation microenvironment was found using conventional immunohistology (lowest inset) by staining for Mena (with anti-Mena) in tumor cells (red), anti-CD68 as a macrophage-specific marker, and anti-CD31 as an endothelial cell marker. A perivascular macrophage is shown in brown touching a Mena-expressing tumor cell that defines this structure as TMEM (Robinson et al. 2009).
Methods for collecting the invasive subpopulations of tumor cells and macrophages during invasion were invented (which we discuss below in the section Visualization and Capture of the Invasive Population of Cells in Primary Tumors) based on these observations and coupled to expression profiling of small numbers of invasive cells to reveal the identities of the genes correlated with the survival, adjuvant resistance, and chemotaxis of invasive cancer cells inside living tumors (Wang et al. 2003, 2004, 2006, 2007; Goswami et al. 2004; Xue et al. 2006; Goswami et al. 2009). These genes fall into well-defined pathways and are coordinately regulated in metastatic tumor cells (Condeelis et al. 2005; Wang et al. 2005, 2007). These pathways are collectively called the Invasion Signature. A major insight to emerge from these studies was that the motility pathways of the Invasion Signature define the mechanisms for tumor-cell migration in vivo (Condeelis et al. 2005).
Molecular markers derived from the mouse Invasion Signature have been extended to human breast tumors so as to determine if any of the master genes of the pathways of the Invasion Signature have a prognostic value. A gene that is strongly up-regulated in invasive tumor cells collected from rat, mouse, and human mammary tumors is the gene for Mena (Wang et al. 2007; Goswami et al. 2009), an Ena/VASP protein. Ena/VASP proteins regulate cell motility by controlling the geometry of assembling actin networks (Krause et al. 2003). Mena appears in the Invasion Signature as regulating signals from the EGF receptor through PI3K and the Rho family to capping protein to increase the lifetime of barbed ends produced by the cofilin and N-WASP pathways.
As summarized above, multiphoton-based intravital imaging had demonstrated that invasive carcinoma cells in mouse and rat mammary tumors intravasate when associated with perivascular macrophages, thereby identifying a tumor microenvironment of metastasis as an anatomical structure in tumors (Fig. 3) (Wyckoff et al. 2007). Therefore, we defined the tripartite arrangement, by triple immunohistochemistry, of an invasive carcinoma cell (marked by Mena overexpression), a macrophage, and an endothelial cell as TMEM (tumor microenvironment of metastasis). TMEM has been identified in human breast tumors using this triple immunohistochemistry technique (Fig. 3) (Robinson et al. 2009). In a retrospective study, we determined that TMEM density in human breast carcinoma samples predicts the development of systemic hematogenous metastases. A case–control study of 30 patients who developed metastatic breast cancer and 30 patients without metastatic disease was performed. Cases were matched to controls based on currently used prognostic criteria. Paraffin-embedded primary breast-cancer samples were stained using the triple immunohistochemical method, allowing simultaneous identification of carcinoma cells, macrophages, and endothelial cells (TMEM). Two pathologists, blinded to outcome, evaluated the number of TMEMs per 20 high-power fields. No association was seen between TMEM density and tumor size, lymph-node metastasis, lymphovascular invasion, or hormone-receptor status. TMEM density was greater in the group of patients who developed systemic metastases compared with the patients with only localized breast cancer (mean = 112 vs. 55, respectively, p = 0.00006). For every increase in TMEM of 10, the risk of systemic metastasis increased by 90% (OR, 1.9; 95% confidence interval [CI] = 1.1–3.4). This study indicates that TMEM is a useful and novel prognostic marker for hematogenous metastasis of human breast tumors (Robinson et al. 2009). This work also illustrates the power of combining multiphoton imaging with mouse models of breast cancer in the development of new insights into metastasis and the microenvironments essential to dissemination of tumor cells in vivo and the identification of markers for both.
ANIMAL MODELS FOR MULTIPHOTON IMAGING: CELL LINES
Transgenic models are ideal for the study of tumors with histology similar to that present in human patients. However, the preparation and maintenance of transgenic models are extremely expensive and time consuming. A relatively quick and inexpensive method for assessing tumor-cell phenotype in vivo is the use of cell lines that express fluorescent proteins. We have used the MTLn3 and MTC cells isolated from a rat mammary tumor as amatched pair of metastatic and nonmetastatic cell types, respectively. It should be noted that the methods for use of cell lines in multiphoton imaging of tumor-cell behavior in vivo described here apply to all cell lines tested to date.
MTLn3-GFP cells were created by Lipofectamine transfection of parental MTLn3 cells with pEGFP-N1 (Clontech) (Farina et al. 1998). MTC-GFP cells were created using retroviral transfection of parental MTCcells with a retroviral GFP-expression construct. The GFP sequence was excised from pEGFPN1 using NsiI and EcoRI and subcloned into the BamHI/EcoRI site of pLXSN. This construct was then transfected into Phoenix cells (created by Gary Nolan of Stanford University) using standard methods (Miller and Rosman 1989; Kinsella and Nolan 1996) and allowed to grow for 24–48 h. The supernatant was collected, filtered, and spun for 5 min at 1000 rpm, and 1 mL was overlaid on a confluent plate of MTC cells. Positive clones were selected by neomycin selection and GFP fluorescence. Stable cells were cultured as parental cells (Segall et al. 1996). Cell growth rate and morphology of transfected cells were determined to be the same as parental cells, and fluorescence was shown to remain constant for 30 passages. Extensive histopathology studies for the MTLn3-GFP cells (Farina et al. 1998) and the MTC-GFP cells were carried out to confirm that their metastatic potentials were similar to the parental cell lines, and this held true for all cell lines expressing members of the GFP family. To create mammary tumors, 1 × 106 cells were injected under the second nipple anterior from the tail of a Fischer 344 rat and allowed to grow for 3.5 wk.
IMAGING ANIMAL MODELS WITH MAMMARY TUMORS
The methods used for handling animals during multiphoton imaging are identical for both transgenic mouse tumors and tumors derived from the injection of cell lines. Rats and mice with mammary tumors are placed under anesthesia with 5% isofluorane and maintained for the course of the imaging session between 0.5% and 2.5% isofluorane to control the rate of breathing. The tumor is exposed by creating a skin flap. This is done by inserting a scalpel between the epidermis and dermis to peel back the epidermis to expose and optically couple the dermis to the objective. The mammary tumor lies below the dermis and is readily imaged, as are cells within the ECM of the dermis. If done properly, the extent of tissue damage is equivalent to that caused by a tattoo and produces minimal inflammation and no disruption of the blood supply and microvasculature of the tumor. During imaging, we have found that fat and dense ECM can be detriments to a quality image. In these cases, the surgery performed on a tumor to remove overlaying fat must be carefully carried out so as not to cut through the vasculature across the top of the tumor and so as not to disrupt the cells in the tumor to avoid destroying the microenvironment. This precaution is particularly important in highly necrotic tumors, which can burst during surgery.
Once prepared for imaging, the animal is placed onto the stage of amultiphoton microscope, and, using 20× 0.95-numerical-aperture (NA), 40× 0.80-NA, or 60× 1.10-NA water-immersion objectives (Fig. 4), is imaged in time lapse, with a single image being captured by a line speed of 50 lps being taken every 2 min with a stack of three to five frame averages in each slice being taken every minute (differences in multiphoton systems are discussed below in the section on Multiphoton Microscopy). On average, each field is imaged for 20–30 min, and each animal can be maintained on the microscope for 3–4 h without special heating or ventilation equipment. Maintaining breathing without drift or movement of the sample is the key to successful intravital imaging. This is achieved by adjusting the level of isofluorane that is administered to keep breathing even and steady while still keeping the animal alive. Imaging areas near the chest cavity is more difficult and means the animal must be kept more deeply sedated. This can affect the length of an imaging session. Most injectible anesthetics, such as ketamine, do not suppress breathing and cannot be monitored or controlled during a long imaging session and are therefore unsuitable for intravital imaging of mammary tumors.
FIGURE 4.
Standard method for imaging a mouse on a multiphoton microscope. A mouse with a GFP-labeled orthotopically injected tumor is shown on the inverted microscope stand (preferred) under isofluorane anesthesia (inset) on a multiphoton microscope. The anesthesia hose and temperature-control box are illustrated.
Transgenic PyMT tumor-bearing mice are allowed to develop tumors for 10–16 wk prior to imaging. In general, other oncogene-driven tumor models progress more slowly, and the optimum time for imaging must be empirically determined and guided by the goals of each experiment.
Multiphoton Microscopy
For imaging samples with fluorophores that can be excited with a standard femtosecond-pulsed laser (e.g., GFP, CFP, or Texas red dextran), we use a turnkey multiphoton system, in our case, the BioRad Radiance 2000 (Fig. 4). In this microscope, the output of a femtosecond-pulsed laser system (Millennia-Tsunami, Newport/Spectra-Physics) passes through a beam-conditioning unit composed of an attenuator, a shutter, and a pair of collimation lenses, which sizes the beam to fill the back-aperture of the objective. From there, the beam is sent on to a galvo pair within the scan head that raster scans the beam within the microscope stand (IX70, Olympus). The microscope has an environmental chamber that can be heated to keep the mouse warm. The temperature in the chamber should not be kept above 30°C, as this will make it harder to keep the mouse under anesthesia (Fig. 4, inset). The fluorescence and second-harmonic signals generated are collected via a dichroic mirror and sent to two photomultiplier-tube (PMT) detectors. The emission filters before the PMTs can be switched to allow detection of CFP (and second harmonic), GFP, and yellow fluorescent protein (YFP). A portion of the emission of rhodamine dextran and Texas red dextran can be detected in the YFP channel. Although the system is limited to only two detectors, we have developed a technique that also allows three channel acquisitions with only two detectors when imaging samples labeled with both CFP and GFP. Because there is a significant bleed of the CFP signal into the GFP channel, adjusting the laser powers and the detector gains to equalize the overlapped signal allows for the separation of the fluors and second-harmonic signals by merging the images into an RGB (red–green–blue) file in the image-processing program ImageJ (http:///rsb.info.nih.gov/ij/). If the CFP/second-harmonic image is put in both the red and the blue channels and the GFP image is placed in the green channel, the CFP will appear white, the GFP will appear green, and the second-harmonic collagen will appear purple (Sahai et al. 2005).
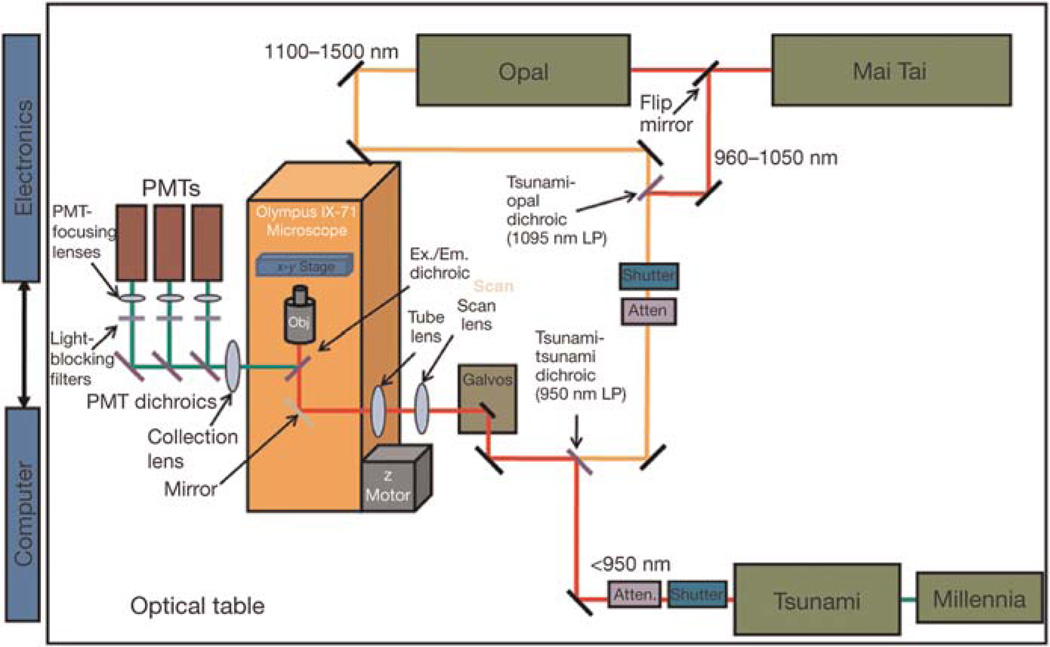
To both extend the range of usable fluorophores and expand the number of available detectors, we have constructed a multiphoton microscope that utilizes two laser-light sources simultaneously (Fig. 5). The first source consists of a standard femtosecond-pulsed laser system (Tsunami–Millennia, Newport/Spectra-Physics) and is used for excitation of fluors in the range of 740–950 nm (e.g., CFP, GFP, YFP). The second source consists of a rapidly tunable femtosecond laser (Mai Tai, Newport/Spectra-Physics) and an optical parametric oscillator (Opal, Newport/Spectra-Physics).
FIGURE 5.
Schematic of the custom multiphoton microscope. By adding an additional light source and additional PMTs, we have increased our ability to do intravital imaging using animals containing multiple fluorescent labels and image endogenous fluorescence by second-harmonic-generated light and by FLIM.
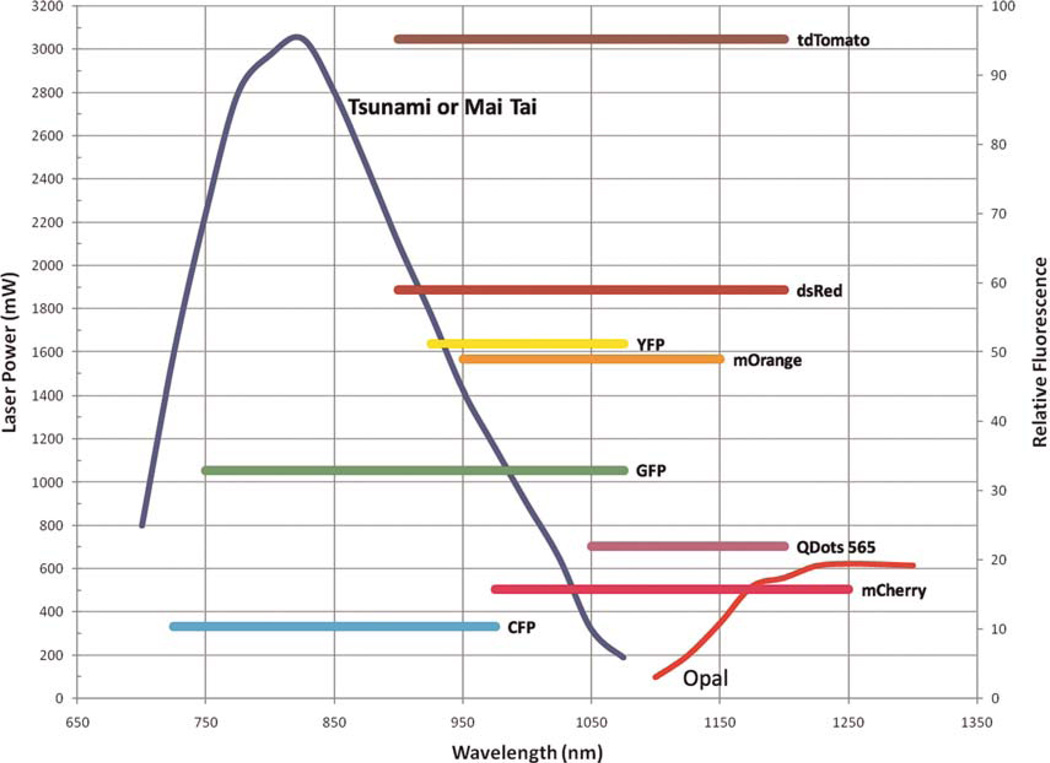
Depending on the combination of fluorophores needed (see Fig. 6), the output from the Mai Tai can be used directly, giving one source with the wavelength range of 960–1050 nm, or it can be tuned to 750 nm and used to pump the optical parametric oscillator where its energy is converted into femtosecond pulses in the range of 1100–1500 nm. A flip mirror and a combining dichroic (1095DRLPXRP, Omega Optical) allow selection between these two possible paths. With either choice of path, the beam is next passed through a shutter and a Pockels-cell attenuator (360–40, Con-Optics). The two beams are combined using a second dichroic (LP02-1064RU-25, Semrock) and sent on to a galvo pair (VM500+, GSI Group). The timing and control signals generating the raster-scan signals for the galvos are created with custom software and a digital-to-analog input/output board (NI PXI-6713, National Instruments). Three nondescanned (2X H7422P-40 module and 1X HC 125-02, Hamamatsu) detectors collect the fluorescence light, and their signals are captured and digitized with two data acquisition boards (PXI-6115, National Instruments). Custom software written in the LabVIEW (National Instruments) programming language controls the entire system from one interface and allows several types of imaging modalities including: time series, z stacks, time lapses, and combinations thereof. Future expansion of the system is simplified, as all electronics and software have been designed to accommodate eight simultaneous acquisition channels. These extra imaging channels will allow simultaneous detection of up to four different cell types, two second-harmonic signals, and two reflectance confocal channels in the same tumor, with the goal of expanding our ability to observe multiple tumor cell–stromal cell interactions and parental cells with multiple mutant tumor cells and/or mutant macrophages in the same tumor (see Fig. 6 for fluorophores that can be used in this system). Tomato is particularly promising as a bright probe, as it has a highly efficient two-photon cross section that increases its fluorescent output (Fig. 7). In addition, imaging of tumor cells in live mammary tumors that express Tomato indicates improved penetration for excitation and emission, allowing imaging to much greater depths (1000 μm) (Fig. 7B). In addition, two of these new detectors are gated, allowing fluorescence lifetime-imaging microscopy (FLIM) of intrinsic fluors, which are present in human tissues and do not contain expression of GFP-like molecules (e.g., NADH [nicotinamide adenine dinucleotide plus hydrogen]) (Bird et al. 2004). This capability will support the multiphoton imaging of human tumor cells and stromal cells in human tissue heterotransplanted into mice, because differences in FLIM between different cell types can be pseudocolored to create images similar to those generated using GFP-expressing cells (Bird et al. 2005).
FIGURE 6.
The multiple fluorescent proteins and injectable agents that can be imaged using the combination of lasers shown in Figure 5. Previously, we were only able to image fluors excitable from incident 760–980-nm light. With the addition of an optical parametric oscillator, we have increased our ability to image fluors out to 1300 nm. The chromophores that can be used are illustrated by comparing the power spectrum of the lasers listed in Figure 5 (axis = laser power) with the multiphoton efficiency of excitation (axis = relative fluorescence). This analysis indicates that Tomato > GFP > CFP and that all can be efficiently imaged.
FIGURE 7.
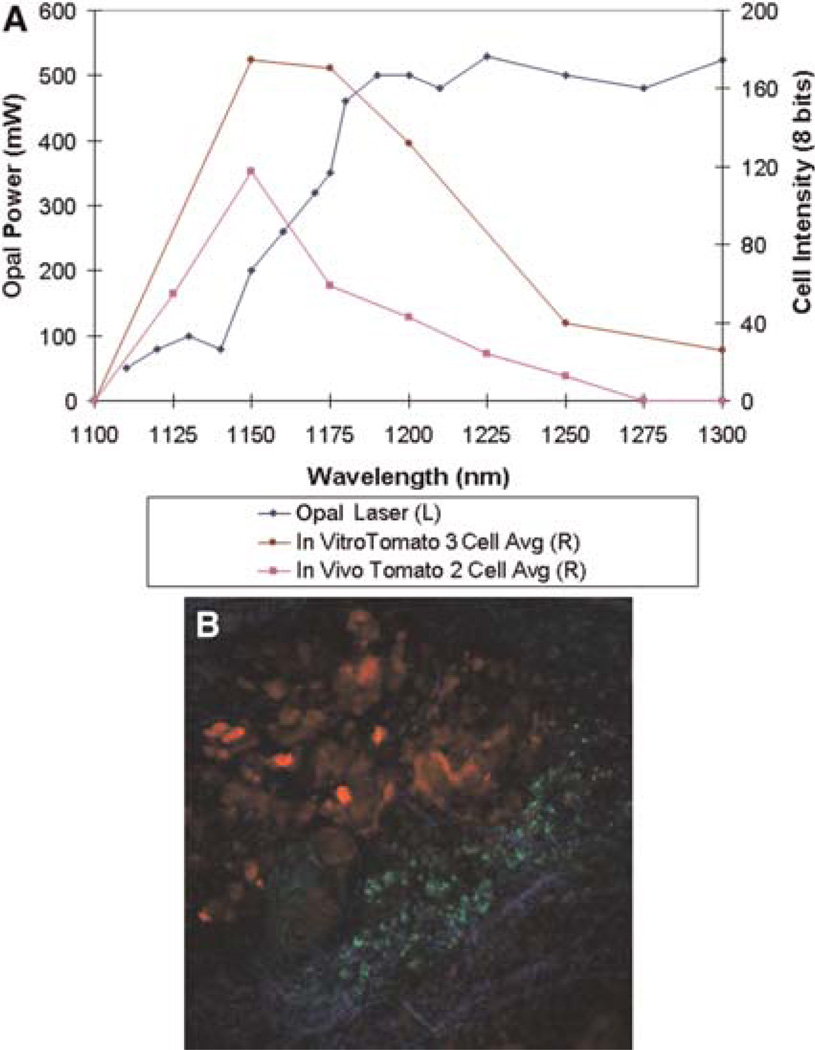
Tomato is a bright far-red chromophore for use in multiphoton imaging. (A) Cells both in culture and in living tumors were imaged using the Opal from 1100 to 1300 nm. Over the power spectrum of the Opal, the maximum intensity of emitted fluorescence (multiphoton efficiency) occurs at ~1150 nm of excitation for cells in vitro and in vivo. L and R refer to left and right axes, respectively. (B) Tomato-labeled MTLn3 cells (red) orthotopically injected into a Scid mouse mammary fat pad are seen imaged using our combined multiphoton/optical parametric oscillator microscope. Macrophages (green) are seen after phagocytosis of Oregon Green dextran. Collagen (blue) is visualized as second-harmonic-generated polarized light.
Vasculature Visualization
For visualizing blood vessels, a key landmark for metastasis in mammary tumors, 200 μL of rhodamine dextran (molecular mass: 2 MDa, Sigma-Aldrich) at 20 mg/mL in Dulbecco’s PBS (phosphate-buffered saline) is injected into the tail vein of the rat or mouse after anesthesia but before surgery. The vasculature in the tumor is then visualized using the red filter in the multiphoton microscope. To image macrophages with Texas red dextran, the animals are injected with 70 kDa of Texas red dextran (Molecular Probes, Invitrogen) as described above and allowed to sit for 2 h postinjection to allow the macrophages to take up the dextran by phagocytosis (see top two insets in Fig. 3) (Farina et al. 1998; Wang et al. 2002; Wyckoff et al. 2007). Other injectable dyes can be used for marking blood vessels, including quantum dots, fluorescently tagged BSA (bovine serum albumin), and fluorescently labeled lectins. Quantum dots can be excited at a single wavelength, but different ones emit at different wavelengths. We have found though that quantum dots tend to aggregate in the vessels and are more variable from lot to lot and so are not as effective as the labeled proteins in delineating vessels.
Imaging Window and Photoswitching
Tracking of individual cells for several hours via the skin-flap method has revealed the existence of tumor-cell invasion and intravasation microenvironments. However, to quantify these behaviors and to determine the physical size of these tumor microenvironments, tracking for longer time periods is required. Skin-flap surgery is usually terminal and limited to a single-imaging session. On the other hand, another widely used animal-imaging approach, which allows several imaging sessions, is the dorsal skinfold chamber (Papenfuss et al. 1979; Lehr et al. 1993). However, this approach introduces limitations of a nonorthotopic environment (breast tumors grown in the skin of the back) and monitoring only very small or two-dimensional (2D) tumors (Dolmans et al. 2002). This is a real problem if one desires to study cell behavior that is specific to the microenvironment of the orthotopic site (e.g., breast tumor in the breast).
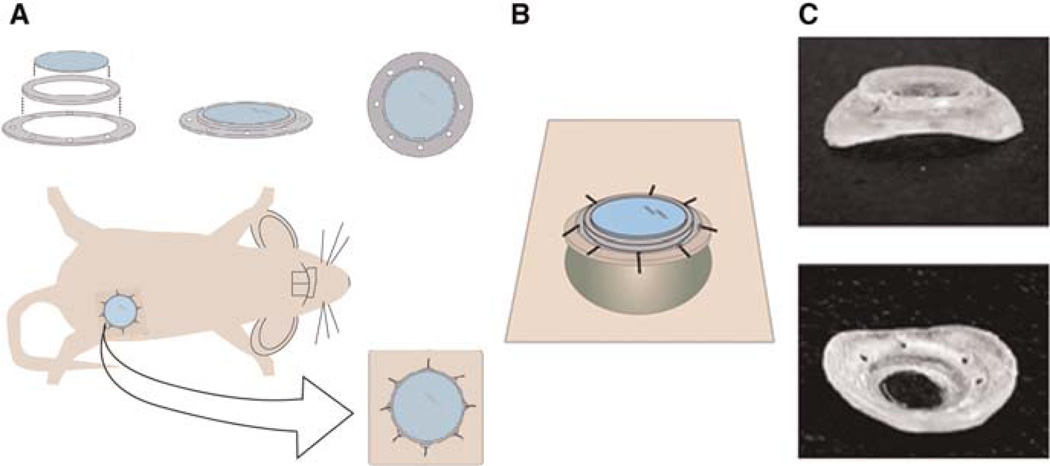
To assess orthotopic breast tumors intravitally at high resolution over multiple-imaging sessions, we developed a mammary-imaging window (MIW) that can be placed on top of the palpable tumor (4–10mmin diameter) or, alternatively, on top of amammary gland of amouse in which we can later introduce tumor cells (Fig. 8). This allows us to use orthotopic mouse xenografts, transgenic mice, or transiently transfected cells injected into the mammary fat pad. The MIW consists of a plastic base with eight suturing holes and a glass coverslip attached on top of the base (Fig. 8A). Use of the glass coverslip assures the optimal working distance and refraction index for high-resolution imaging. Based on the size and the shape of the tumor, the MIW can be assembled to be flat (Fig. 8A) or anatomically curved (Fig. 8B). Implantation of the MIW extends the imaging time from several hours to multiple days (depending on the tumor growth rate, up to 21 d) and is limited solely by the size of the tumor underneath. Tumors with MIW implants do not show a significant difference in rate of tumor growth, macrophage density, angiogenesis, or necrosis compared with tumors without the implant at 1–9 d after the implantation procedure (Kedrin et al. 2008).
FIGURE 8.
MIW components and implantation. The MIW consists of two plastic rings that form a base for a glass coverslip. The mount has holes that facilitate suturing into the skin, whereas the glass coverslip assures the optimal working distance and refraction index for high-resolution imaging. MIW is surgically implanted on top of the mammary gland or the mammary tumor. (A) The view of the window as it would appear sutured into the skin (beige) on top of a growing tumor. (B) The plastic base of the MIW is anatomically shaped to fit on top of the tumor 4–10 mm in diameter (round green mass under the window). (C) Photograph of the MIW from the side and the bottom.
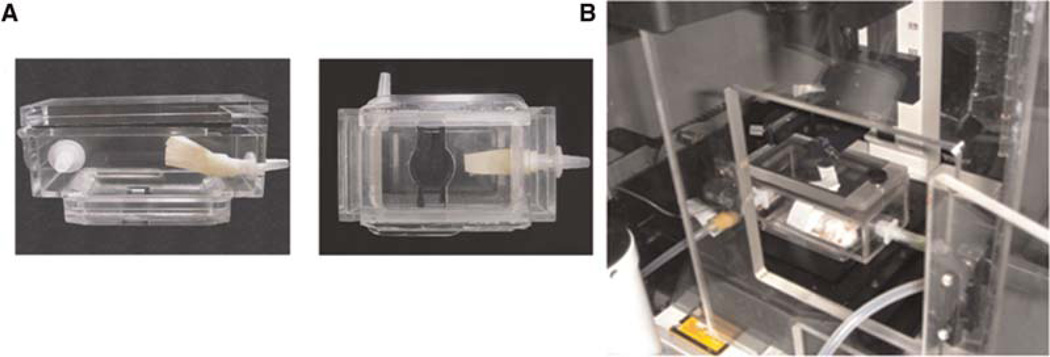
To properly position the MIW on top of the microscope objective and immobilize the animal so that the same field can be repeatedly imaged in all imaging sessions, we place the animal with the MIW implant inside the stereotactic-imaging box (Fig. 9), which also keeps anesthesia levels stable via the inlet (isofluorane/oxygen) and the outlet (vacuum). A flow of anesthesia enters the stereotactic box through the inlet and exits toward the vacuum because of the negative pressure, passing through a carbon filter that removes isofluorane from the mix.
FIGURE 9.
The stereotactic box. (A) View of the stereotactic-imaging box from the side (left) and the bottom (right). The animal is placed inside the box by opening the lid on the top; the imaging window is further securely immobilized between two sliding doors on the bottom of the box. (B) The view of the mouse inside the imaging box. The imaging box is connected to the anesthesia machine (right side) and to the vacuum (front left) and placed inside the environmental chamber built around the microscope stage. The condenser and the slide holder are removed prior to the imaging-box placement.
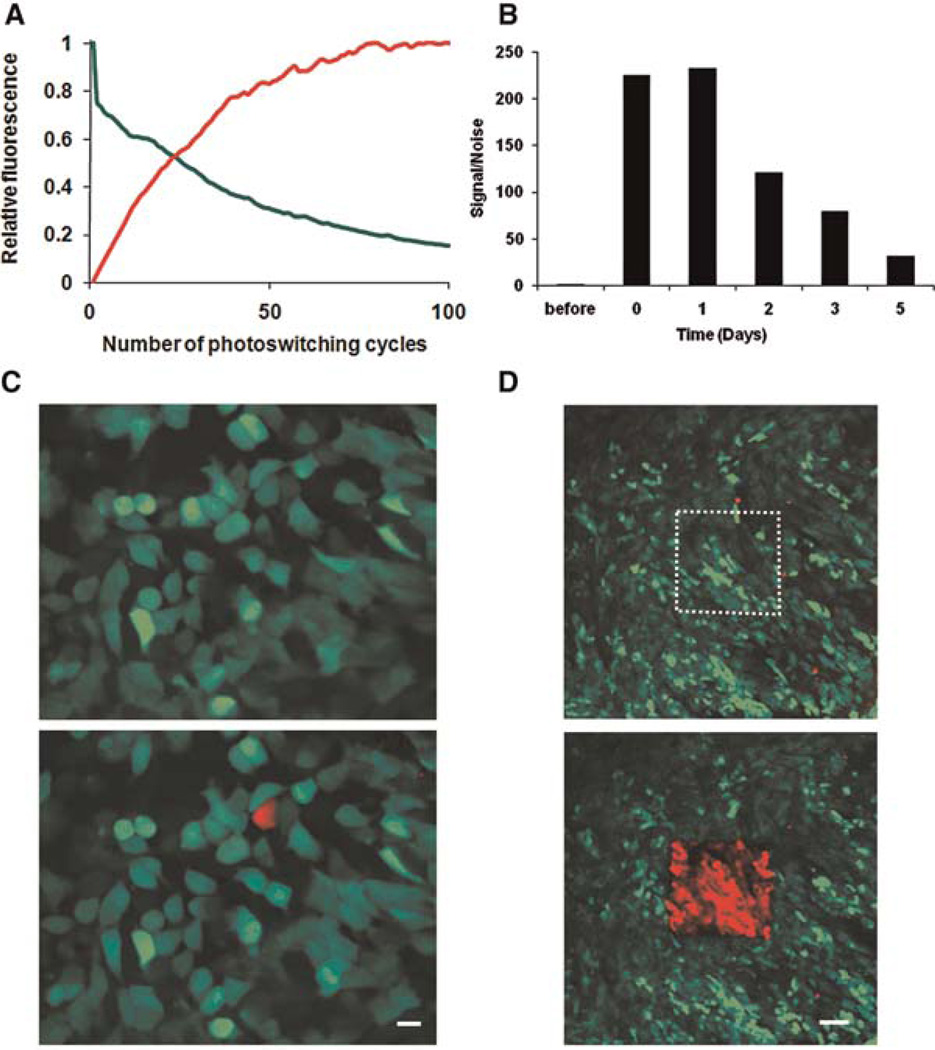
As noted earlier in the article, we are interested in monitoring specific populations of cells over prolonged times and in comparing different microenvironments inside the same animal. However, tissue topology of the tumor changes because of angiogenesis and cell proliferation and migration, limiting the use of fixed reference points such as fluorescent beads attached to the tissue or to the MIW (Entenberg et al. 2006; Bins et al. 2007). To circumvent this problem we used Dendra2, a photo-switchable fluorescent protein (Lukyanov et al. 2005; Gurskaya et al. 2006), which we stably expressed in breast-cancer lines MTLn3 and MDA-MB-231, to directly photomark cells of interest. Dendra2 resembles GFP in its spectrum prior to photoswitching, but exposure to blue light (e.g., 405-nm single photon and 810-nm two photon) can induce an irreversible redshift >150 nm in the excitation and emission spectra of the chromophore, creating an RFP (red fluorescent protein)-like protein (Chudakov et al. 2007). Following the photoswitch, the red fluorescence stably increases up to 250-fold both in vitro (data not shown) and in vivo (Fig. 10A), resulting in red/green contrast of up to 850× and allowing us to track cells marked in this way. Five days after photoswitching, the red fluorescence of the cells photoswitched inside the tumor is still 31-fold higher than the red fluorescence of nonswitched cells, which enables us to recognize the highlighted cells in vivo for extended times after the photoswitch (Fig. 10B).
FIGURE 10.
Speed, stability, and precision of photoswitching in vivo. (A) Average increase in red and decrease in green signal in Dendra2-MTLn3 cells upon photoswitching, as measured in the region of interest from panel D. The values are normalized to the highest fluorescent level in red and the initial fluorescent level in green. We determined the ideal length of the photoswitching (reaching the plateau in fluorescence) for this region to be 126 sec (i.e., 70 cycles). (B) Cells within Dendra2-MTLn3 tumors were photoswitched through the MIW, and the red fluorescence was quantified before photoswitching, immediately after (0 d) photoswitching, and over the 5 subsequent days. The values were normalized to the red fluorescence level before photoswitching. (C,D) Photoswitching of Dendra2 in vivo can be easily done in regions of interest ranging from one cell (C; scale bar, 10 μm) to hundreds of cells (D; scale bar 75 μm) through the MIW.
Regions of interest containing one to hundreds of cells can be selectively photoswitched and visualized through the MIW (Fig. 10C,D). As cells in the tumor migrate and invade, the distribution of these (red) cells relative to blood vessels and other (green) tumor cells changes over time. To quantify invasion and intravasation within distinct mammary-gland microenvironments, we photoswitch square regions (~300 cells) inside the same orthotopically grown tumor, focusing on regions lacking and containing detectable blood vessels. The location of the photoswitched red cells is determined via acquiring z stacks of the same regions at 0, 6, and 24 h after photoswitching (Fig. 10C,D) (Kedrin et al. 2008).
The combination of photoswitchable proteins with the MIW allows for a quantitative long-term analysis of a distinct group of cells photomarked in the primary tumor and tracked over time without long-term anesthesia. Depending on the experimental goals, we are able to acquire images using the MIW with either single- or multiphoton microscopes. High stability of Dendra2 also enables us to freeze fix the tissues and analyze them without additional labeling. One limitation of the photoswitching approach is that both green and red channels are used to label tumor cells, which, therefore, lowers the number of excitation wavelengths to be used for other components of the microenvironment. The combined use of MIW and photomarking cells to revisit chosen subpopulations of cells is an important capability that allows us to further tackle the link between tumor-cell motility and metastasis in a heterogeneous tumor background.
Visualization and Capture of the Invasive Population of Cells in Primary Tumors
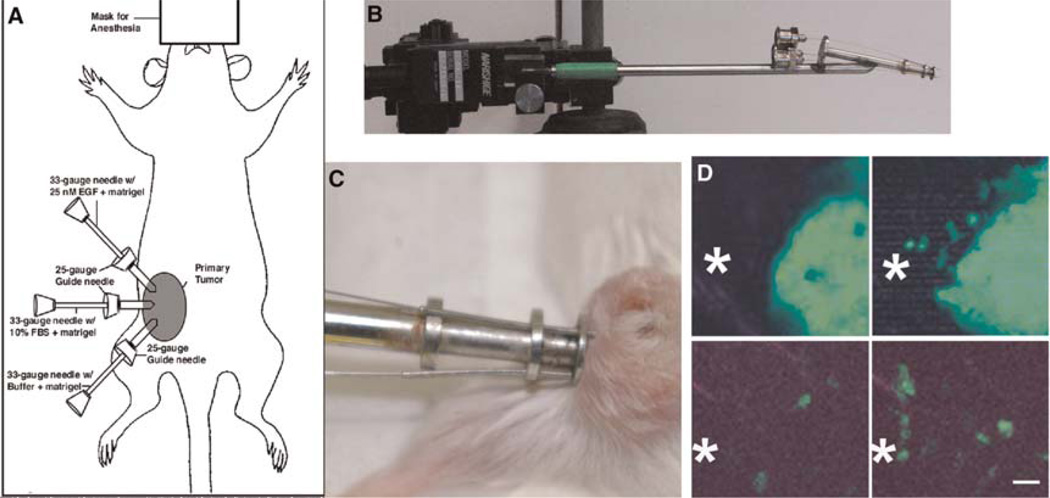
One of the most useful technologies to come from intravital imaging of tumors is the in vivo invasion assay. The in vivo invasion assay was inspired by the multiphoton-imaging observation that tumor cells are spontaneously and chemotactically attracted to blood vessels in the primary mammary tumors of rats and mice (Fig. 11). The in vivo invasion assay is an artificial blood vessel consisting of a 33-gauge needle filled with Matrigel and various growth factors to act as surrogates for blood vessels when inserted into a primary tumor (Wang et al. 2003, 2004, 2006, 2007; Goswami et al. 2004; Wyckoff et al. 2004a,b; Hernandez et al. 2009). The design of the device and its micromanipulator-controlled positioning system allow one to test for growth factors that are effective at attracting invasive tumor cells inside the primary tumor and assessing the stromal cells that are comigratory with invasive tumor cells in vivo. The in vivo invasion assay has been used successfully to collect the invasive tumor cells and accompanying stromal cells from rat and mouse mammary tumors and human heterotransplant tumors of the breast, lung, and head and neck. In mammary tumors, the migration of tumor cells and macrophages into the needles has been imaged by multiphoton microscopy in vivo, demonstrating that tumor and stromal cells actively migrate into the needles consistent with their mimicry of blood vessels (see Fig. 11D).
FIGURE 11.
The in vivo invasion assay can be used to collect invasive tumor cells and accompanying stromal cells from living tumors during intravital imaging. (A) A model for the in vivo invasion assay shows the method for collecting cells from living tumors using needles (ID 102 μm) filled with Matrigel and the ligand of interest when placed in the mammary tumor of an anesthetized animal. (B) Holding apparatus for 25-gauge guide needles and 33-gauge experimental needles is shown attached to micromanipulator used to precisely place needles into the mammary tumor. (C) Living tumor in anesthetized mouse is shown with needles inserted. The needles are left in the tumor for up to 4 h. (D) Movement of tumor cells (top, WAP-Cre/CAG-CAT-EGFP/MMTV-PyMT tumor) and macrophages (bottom, MMTV-PyMT/lys-GFPKi) toward EGF-containing needles. The approximate opening of the collection needle is shown in the field as *. Each image is a 50-μm z projection from a time-lapse series. Images on the right were recorded 90 min after images on the left. Scale bar, 25 μm.
By using the in vivo invasion assay, we can collect the subpopulation of invasive tumor cells, observed in vivo by imaging, as a live cell population. This collection of cells can be subjected to expression profiling to correlate cell behavior in vivo with gene expression (Wang et al. 2006, 2007). A detailed protocol for the preparation and insertion of collection needles is described in The In Vivo Invasion Assay: Preparation and Handling of Collection Needles (Wyckoff et al. 2011).
GENE DISCOVERY BASED ON BEHAVIORAL ANALYSIS
Correlated multiphoton imaging and collection of live cells into needles in the in vivo invasion assay demonstrate that the collection of migratory cells from the primary tumor is efficient enough to provide a window on the behavior and properties of the invasive cells within the primary tumor (Wang et al. 2004). Multiphoton imaging is performed in combination with the in vivo invasion assay to confirm that the tumor cells and their accompanying stromal cells are actively invading the needles of the in vivo invasion assay using cell motility and that cell collection using this assay is not a passive result from insertion of the collection needles or some other artifact resulting from the methodology. The use of imaging to confirm the presence of invasive tumor cells moving into the collection needles has been described in detail previously (Wang et al. 2004). The combination of genetically derived animal models of metastasis with multiphoton imaging and cell collection represents a truly unique and innovative approach to the study of invasion and metastasis in vivo. This combined technology has allowed the characterization of gene-expression patterns of invasive tumor cells isolated at the moment of invasion from the primary tumor (Wang et al. 2003, 2004, 2005, 2006, 2007; Condeelis et al. 2005; Goswami et al. 2005, 2009). By further comparing the gene-expression patterns of invasive tumor cells collected by invasion into the needles of the in vivo invasion assay with those of tumor cells obtained from the whole primary tumor, the blood, and whole metastatic tumors, genes that contribute uniquely to the invasive process in tumor cells have been identified. This set or pathway of genes is called the Invasion Signature (described in the section Animal Models for Multiphoton Imaging: Transgenic Mice), and it has supplied a number of biomarkers useful in prognosis (Robinson et al. 2009). Using these approaches, it has been possible to demonstrate that the changes in gene expression of the Invasion Signature of rats and mice fall into two categories: transient and stable (Goswami et al. 2009). Some differential expression is lost when the invasive tumor cells are cultured as primary cells and also spontaneously in vivo during later stages of metastasis, whereas other genes of the Invasion Signature retain their differential expression during culture and during progression to later stages of metastasis. The nature of this mechanism is currently being evaluated in mice and has been useful in identifying Mena as a biomarker for metastasis in humans (Robinson et al. 2009).
MEASUREMENT OF CELL BEHAVIOR IN VIVO
Cell motility, polarity, intravasation, and adhesion are visualized by time-lapse multiphoton microscopy by taking an image at 2-min intervals over a period of at least 30 min. Currently, each image requires 10 sec for collection to provide good spatial resolution for cells moving 10 μm/min or less. The images are then assembled into four-dimensional (4D) movies in ImageJ. Animation of the movies allows for the detection of cell motility, intravasation, and cell protrusion. From these movies, we have used manual tracing to determine the number of cells translocating, the velocity and distance traveled by cells, polarization and orientation of cells toward blood vessels, protrusive movement of pseudopods and invadopods, and the orientation and movement of cells around landmarks such as ECM and macrophages (Farina et al. 1998; Wang et al. 2002, 2007; Sidani et al. 2006; Wyckoff et al. 2007).
Cell polarization, especially orientation toward blood vessels, is determined by observing the shape of the cells within the field. A polarized cell is characterized as a cell with a distinct leading edge and a length/width ratio >1.5 (Sidani et al. 2007). The polarization of cells around blood vessels is used to determine if cells are oriented toward the vessels, compared with those displaying random orientation. Percent orientation was determined as the percentage of blood vessels per 200-μm imaging field with four or more directly adjacent cells polarized toward the vessel. Percent orientation was corrected for randomly polarized fields of cells subtracting from the above value the percentage of blood vessels with four or fewer cells polarized toward the vessel (Sidani et al. 2006; Wang et al. 2007).
Host cells can be imaged in three ways. First, they can be imaged as shadows crawling on top of fluorescent carcinoma cells (Farina et al. 1998). Second, macrophages can be imaged by loading through intravenous injection with rhodamine-labeled dextran, which they then phagocytose. Finally, specific cell types can be imaged in transgenic mice by expression of GFP by cell-type-specific promoters—for example, using the lys or CSF-1 promoter to drive GFP expression in macrophages as described in the section Animal Models for Multiphoton Imaging: Transgenic Mice (Faust et al. 2000; Wyckoff et al. 2004a,b).
Because of the ability to image ECM fibers by second-harmonic light scattering, cell–matrix interactions can also be inferred. These interactions include cell motility along matrix fibers and adhesion (Wang et al. 2002; Condeelis and Segall 2003). To image adhesion, one compares amovie showing just the matrix channel together with a movie of the combined cell and matrix channels. Sites of cell colocalization with the matrix that correlate with shadows that appear on the matrix are counted as adhesion sites. Adhesion cannot be directly quantitated; however, differences in cell adhesion can be determined by differences in intensity of second-harmonic signals from collagen fibers between locations where cells are in contact compared with those observed in cell-free regions. Cell locomotion that tracks exactly along collagen fibers can be scored as ameasure of cell adhesion (Wang et al. 2002). Comparative matrix density between differing tumor volumes is determined by calculating the pixel intensity from a reconstructed z series of just the matrix channel (Wang et al. 2002).
More recently, an ImageJ plug-in (MTrackJ, Biomedical Imaging Group Rotterdam) has been used for the determination of such variables as velocity, total path length, and net path length. From this information, such variables as persistence and directionality can be calculated. Further, this plug-in allows for the delineation of the cell tracks that can then be used to illustrate the cell-movement paths (Egeblad et al. 2008). Other software such as Volocity (Improvision) is used for centroid plots. Perimeter plots of shape changes of individual cells are performed using Adobe ImageReady CS2 (Cvejic et al. 2008) or an equivalent (e.g., Adobe Photoshop C3). High-resolution 4D reconstructions are accomplished using rendering software (Imaris; Bitplane).
ACKNOWLEDGMENTS
We thank Mazen Sidani, Erik Sahai, Jacco van Rheenen, and Dimitri Kedrin for their contributions to the technology of intravital imaging discussed in this article. We also thank the staff of the Analytical Imaging Facility at Albert Einstein College of Medicine for their help with this article. This work was supported by grants from the NIH including CA100324, CA113395, and CA126511 and from the US Department of Defense W81XWH0501405.
Footnotes
WWW RESOURCE
http://www.einstein.yu.edu/biophotonics/ Advanced Optical Microscope used in studies described in this article are part of the Gruss Lipper Biophotonics Center.
REFERENCES
- Ahmed F, Wyckoff J, Lin EY, Wang W, Wang Y, Hennighausen L, Miyazaki J, Jones J, Pollard JW, Condeelis JS, et al. GFP expression in the mammary gland for imaging of mammary tumor cells in transgenic mice. Cancer Res. 2002;62:7166–7169. [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Bins AD, van Rheenen J, Jalink K, Halstead JR, Divecha N, Spencer DM, Haanen JB, Schumacher TN. Intravital imaging of fluorescent markers and FRET probes by DNA tattooing. BMC Biotechnol. 2007;7:2. doi: 10.1186/1472-6750-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DK, Eliceiri KW, Fan CH, White JG. Simultaneous twophoton spectral and lifetime fluorescence microscopy. Appl Opt. 2004;43:5173–5182. doi: 10.1364/ao.43.005173. [DOI] [PubMed] [Google Scholar]
- Bird D, Yan L, Vrotsos K, Eliceiri K, Keely PJ, White JG, Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65:8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- Böttinger EP, Jakubczak JL, Haines DC, Bagnall K, Wakefield LM. Transgenic mice overexpressing a dominant-negative mutant type II transforming growth factor <gb> receptor show enhanced tumorigenesis in the mammary gland and lung in response to the carcinogen 7,12-dimethylbenz-[a]-anthracene. Cancer Res. 1997;57:5564–5570. [PubMed] [Google Scholar]
- Butler TP, Gullino PM. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 1975;35:512–516. [PubMed] [Google Scholar]
- Campagnola PJ, Clark HA, Mohler WA, Lewis A, Loew LM. Secondharmonic imaging microscopy of living cells. J Biomed Opt. 2001;6:277–286. doi: 10.1117/1.1383294. [DOI] [PubMed] [Google Scholar]
- Carter D. Interpretation of breast biopsies. New York: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75:2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudakov DM, Lukyanov S, Lukyanov KA. Tracking intracellular protein movements using photoswitchable fluorescent proteins PS-CFP2 and Dendra2. Nat Protoc. 2007;2:2024–2032. doi: 10.1038/nprot.2007.291. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Pollard J. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3:921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- Condeelis JS, Wyckoff J, Segall JE. Imaging of cancer invasion and metastasis using green fluorescent protein. Eur J Cancer. 2000;36:1671–1680. doi: 10.1016/s0959-8049(00)00155-6. [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH, Segall JE. THE GREAT ESCAPE: When cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- Cvejic A, Hall C, Bak-Maier M, Flores MV, Crosier P, Redd MJ, Martin P. Analysis of WASp function during the wound inflammatory response-Live-imaging studies in zebrafish larvae. J Cell Sci. 2008;121:3196–3206. doi: 10.1242/jcs.032235. [DOI] [PubMed] [Google Scholar]
- Dolmans DE, Kadambi A, Hill JS, Waters CA, Robinson BC, Walker JP, Fukumura D, Jain RK. Vascular accumulation of a novel photosensitizer, MV6401, causes selective thrombosis in tumor vessels after photodynamic therapy. Cancer Res. 2002;62:2151–2156. [PubMed] [Google Scholar]
- Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, Peeters G, Krummel MF, Werb Z. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Models Mech. 2008;1:155–167. doi: 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancerIThe value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Entenberg D, Aranda I, Li Y, Toledo-Crow R, Schaer D, Li Y. Multimodal microscopy of immune cells and melanoma for longitudinal studies. Proc SPIE. 2006;6081:62–73. [Google Scholar]
- Farina KL, Wyckoff JB, Rivera J, Lee H, Segall JE, Condeelis JS, Jones JG. Cell motility of tumor cells visualized in living intact primary tumors using green fluorescent protein. Cancer Res. 1998;58:2528–2532. [PubMed] [Google Scholar]
- Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- Glaves D. Detection of circulating metastatic cells. Prog Clin Biol Res. 1986;212:151–167. [PubMed] [Google Scholar]
- Goswami S, Wang W, Wyckoff JB, Condeelis JS. Breast cancer cells isolated by chemotaxis from primary tumors show increased survival and resistance to chemotherapy. Cancer Res. 2004;64:7664–7667. doi: 10.1158/0008-5472.CAN-04-2027. [DOI] [PubMed] [Google Scholar]
- Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- Goswami S, Philippar U, Sun D, Patsialou A, Avraham J, Wang W, Di Modugno F, Nistico P, Bertier FB, Condeelis JS. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin Exp Metastasis. 2009;26:153–159. doi: 10.1007/s10585-008-9225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzburg WH, Salmons B, Zimmermann B, Muller M, Erfle V, Brem G. A mammary-specific promoter directs expression of growth hormone not only to the mammary gland, but also to Bergman glia cells in transgenic mice. Mol Endocrinol. 1991;5:123–133. doi: 10.1210/mend-5-1-123. [DOI] [PubMed] [Google Scholar]
- Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Hernandez L, Smirnova T, Wyckoff J, Condeelis J, Segall JE. In vivo assay for tumor cell invasion. Methods Mol Biol. 2009;571:227–238. doi: 10.1007/978-1-60761-198-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, van Rheenen J, Hernandez L, Condeelis J, Segall JE. Cell motility and cytoskeletal regulation in invasion and metastasis. J Mammary Gland Biol Neoplasia. 2007;12:143–152. doi: 10.1007/s10911-007-9046-4. [DOI] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5:1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- Lehr HA, Leunig M, Menger MD, Nolte D, Messmer K. Dorsal skinfold chamber technique for intravital microscopy in nude mice. Am J Pathol. 1993;143:1055–1062. [PMC free article] [PubMed] [Google Scholar]
- Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Kleinerman J, Saidel GM. Quantitative relationships of intravascular tumor cells, tumor vessels, and pulmonary metastases following tumor implantation. Cancer Res. 1974;34:997–1004. [PubMed] [Google Scholar]
- Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- Muller WJ, Ho J, Siegel PM. Oncogenic activation of Neu/ErbB-2 in a transgenic mouse model for breast cancer. Biochem Soc Symp. 1998;63:149–157. [PubMed] [Google Scholar]
- Papenfuss HD, Gross JF, Intaglietta M, Treese FA. A transparent access chamber for the rat dorsal skin fold. Microvasc Res. 1979;18:311–318. doi: 10.1016/0026-2862(79)90039-6. [DOI] [PubMed] [Google Scholar]
- Price JT, Bonovich MT, Kohn EC. The biochemistry of cancer dissemination. Crit Rev Biochem Mol Biol. 1997;32:175–253. doi: 10.3109/10409239709082573. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Robinson B, Sica G, Liu Y-F, Rohan T, Gertler F, Condeelis J, Jones J. Tumor microenvironment of metastasis (TMEM) in human breast carcinoma: A potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15:2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Wyckoff J, Philippar U, Segall JE, Gertler F, Condeelis J. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasmono R, Oceandy D, Pollard J, Tong W, Pavli P, Wainwright B, Ostrowski M, Himes S, Hume D. A macrophage colonystimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Segall JE, Tyerech S, Boselli L, Masseling S, Helft J, Chan A, Jones J, Condeelis J. EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin Exp Metastasis. 1996;14:61–72. doi: 10.1007/BF00157687. [DOI] [PubMed] [Google Scholar]
- Sidani M, Wyckoff K, Xue C, Segall JE, Condeelis J. Probing the microenvironment of mammary tumors using multiphoton microscopy. J Mammary Gland Biol Neosplasia. 2006;11:151–163. doi: 10.1007/s10911-006-9021-5. [DOI] [PubMed] [Google Scholar]
- Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, Wang W, Kuhl S, El-Sibai M, Backer EM, et al. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol. 2007;179:777–791. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Ryan ED, Cardiff RD, Muller WJ. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: Implications for human breast cancer. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soon L, Braet F, Condeelis J. Moving in the right directionnanoimaging in cancer cell motility and metastasis. Microsc Res Tech. 2007;70:252–257. doi: 10.1002/jemt.20411. [DOI] [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Frohlich VC, Oleynikov Y, Huttelmaier S, Zavadil J, Cermak L, Bottinger EP, Singer RH, White JG, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62:6278–6288. [PubMed] [Google Scholar]
- Wang W, Wyckoff JB, Wang Y, Bottinger EP, Segall JE, Condeelis JS. Gene expression analysis on small numbers of invasive cells collected by chemotaxis from primary mammary tumors of the mouse. BMC Biotechnol. 2003;3:13–25. doi: 10.1186/1472-6750-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Goswami S, Lapidus K, Wells A, Wyckoff J, Sahai E, Singer R, Segall J, Condeelis J. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- Wang W, Goswami S, Sahai E, Wyckoff JB, Segall JE, Condeelis JS. Tumor cells caught in the act of invading: Their strategy for enhanced cell motility. Trends Cell Biol. 2005;15:138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wyckoff J, Wang Y, Goswami S, Sidani M, Condeelis J. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Re. s. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- Webster MA, Muller WJ. Mammary tumorigenesis and metastasis in transgenic mice. Semin Cancer Biol. 1994;5:69–76. [PubMed] [Google Scholar]
- Williams RM, Zipfel WR, Webb WW. Multiphoton microscopy in biological research. Curr Opin Chem Biol. 2001;5:603–608. doi: 10.1016/s1367-5931(00)00241-6. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Jones JG, Condeelis JS, Segall JE. A critical step in metastasis: In vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004a;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Segall J, Condeelis J. Single-cell imaging in animal tumors in vivo. In. In: Goldman RD, Spector DL, editors. Live cell imaging, A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004b. pp. 409–422. [Google Scholar]
- Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCKand myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. The in vivo invasion assay: Preparation and handling of collection needles. Cold Spring Harb Protoc. 2011 doi: 10.1101/pdb.prot065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Wyckoff J, Liang F, Sidani M, Violini S, Tsai KL, Zhang ZY, Sahai E, Condeelis J, Segall JE. Epidermal growth factor receptor overexpression results in increased tumor cell motility in vivo coordinately with enhanced intravasation and metastasis. Cancer Res. 2006;66:192–197. doi: 10.1158/0008-5472.CAN-05-1242. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Pixley F, Condeelis J. Invadopodia and podosomes in tumor invasion. Eur J Cell Biol. 2006;85:213–218. doi: 10.1016/j.ejcb.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Yuan HT, Suri C, Landon DN, Yancopoulos GD, Woolf AS. Angiopoietin-2 is a site-specific factor in differentiation of mouse renal vasculature. J Am Soc Nephrol. 2000;11:1055–1066. doi: 10.1681/ASN.V1161055. [DOI] [PubMed] [Google Scholar]