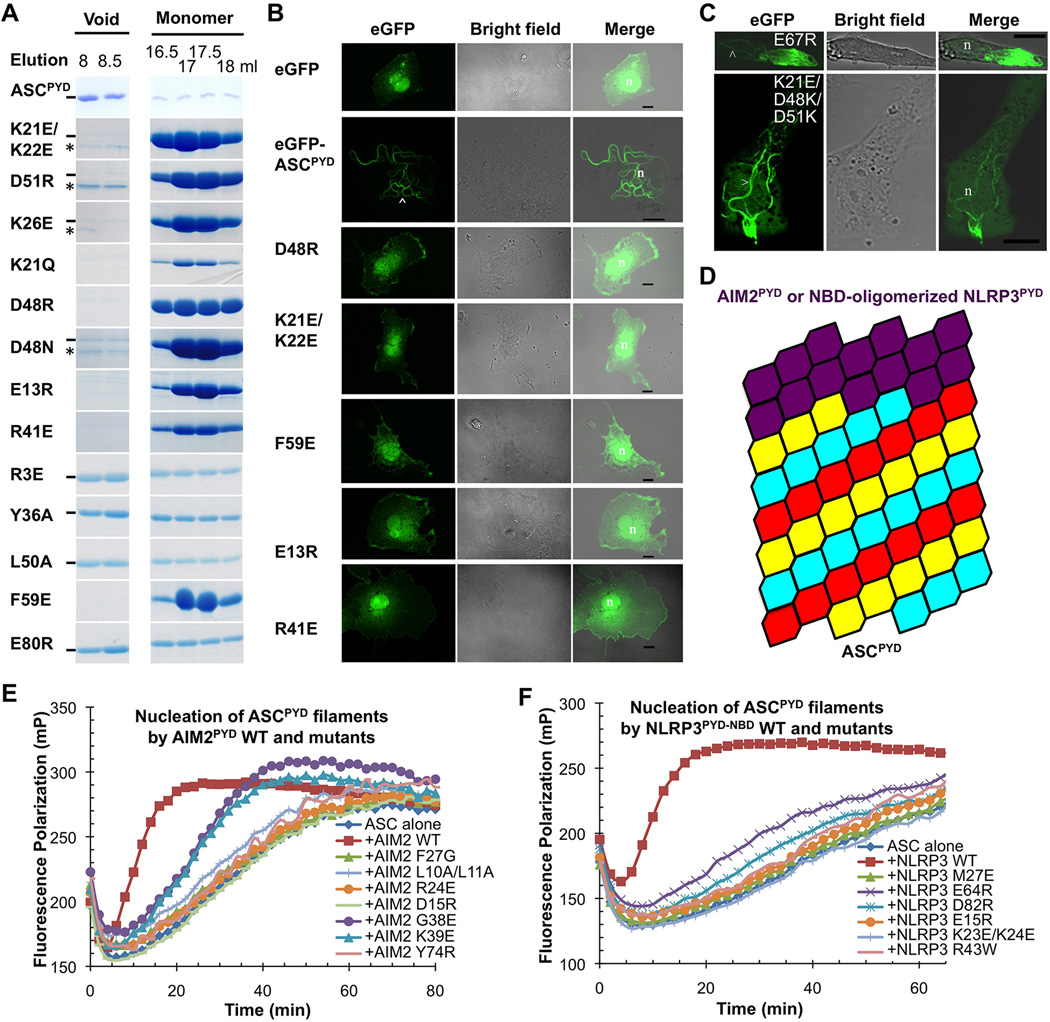
Figure 5. Structure-based Mutations Disrupts ASCPYD Filament Formation, AIM2PYD/ASCPYD Interaction and NLRP3PYD/ASCPYD Interaction in Vitro and in Cells.
A. Size-exclusion chromatography of WT and mutant ASCPYD showing both filamentous (void) and monomeric fractions from a Superdex 200 column. Hyphen denotes ASCPYD and asterisk denotes a contaminant.
B. Morphology of transfected WT and mutant eGFP-tagged ASCPYD constructs visualized by confocal laser scanning microscopy. The arrowhead depicts filaments. n: nucleus; scale bars = 10µm.
C.Morphology of transfected eGFP-tagged ASCPYD visualized by confocal laser scanning microscopy. Top: ASCPYD with charge reversal mutation on a residue outside the filament interface. Bottom: ASCPYD with triple charge reversal mutation that rescued the defectiveness of the single mutants. Arrow heads depict filaments.
D.A schematic model of AIM2PYD/ASCPYD or NLRP3PYD/ASCPYD filaments composed of a top AIM2PYD or NLRP3PYD layer extended by ASCPYD filament body.
E,F. Mutations of conserved interfacial residues on AIM2PYD (E) and NLRP3PYD-NBD (F) reduced or abolished their ability to nucleate ASCPYD filaments.
See also Figure S5.