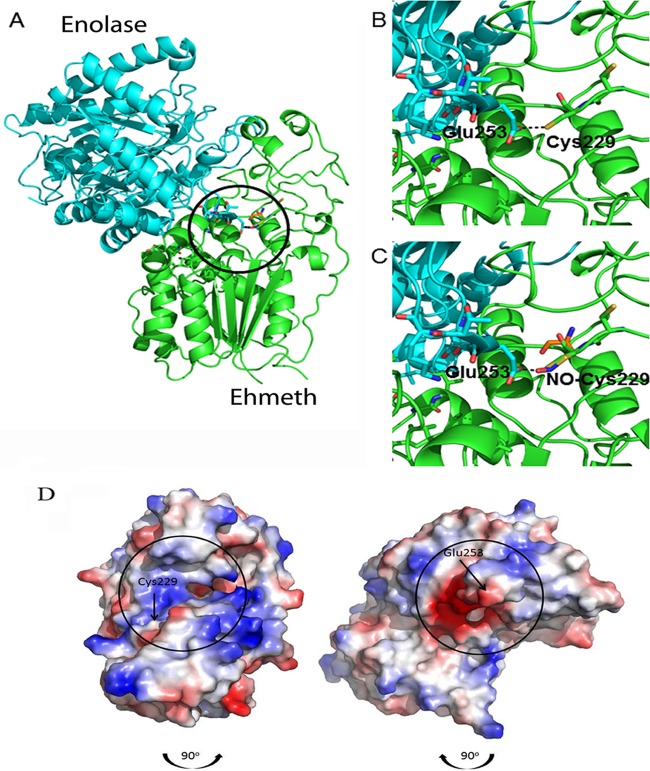
FIG 4.
Molecular modeling of the putative enolase-Ehmeth complex. The atomic coordinates of E. histolytica enolase and Ehmeth proteins (PDB codes 3QTP and 3QV2, respectively) were docked using the Hex 6.2 platform. (A) The overall best docked structure. (B and C) Close-up images of the interaction between the Glu253 residue of enolase and the Cys229 residue of Ehmeth in the native (B) and NO-modified (C) forms. The complex shown in panel A was disassembled by rotating the enolase 90° in a clockwise direction and the Ehmeth 90° in a counterclockwise direction (the black arrows indicate the direction of complex formation). The interaction interfaces of both proteins are indicated by black circles. (D) Vacuum electrostatic potentials were generated using PyMOL in order to illustrate the charge variance of the E. histolytica enolase and Ehmeth proteins, with red and blue indicating negative and positive surfaces, respectively.