Fig. 3.
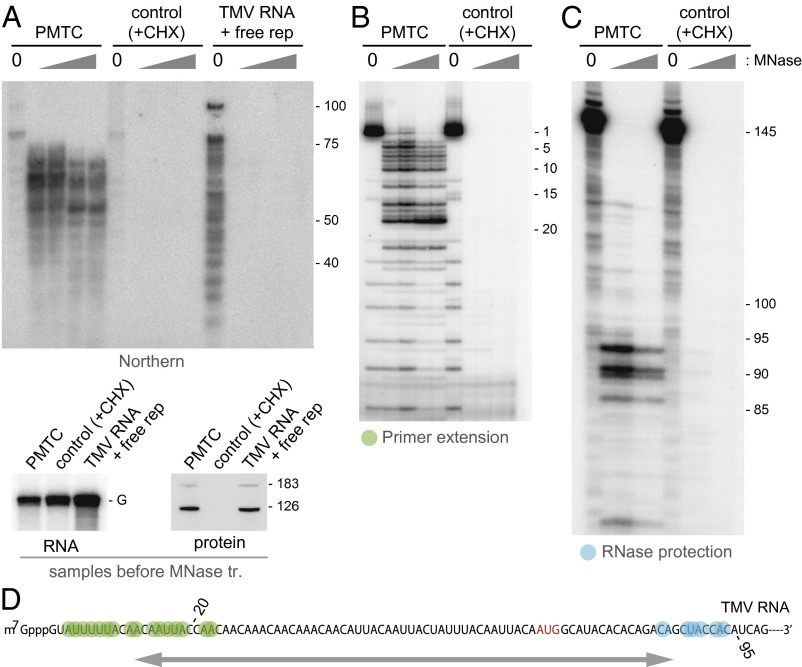
Characterization of TMV RNA fragments generated by MNase treatment of the PMTC. (A) (Upper) Northern hybridization analysis of MNase-resistant RNA fragments. The PMTC fraction, the control SEC fraction, and TMV RNA incubated with fraction no. 95 containing TMV RNA-free replication proteins were treated with 0, 0.05, 0.1, 0.5, or 1 U/μL MNase. RNA was purified from the reaction mixtures, separated by 8 M urea-10% (19:1 bis) PAGE, and analyzed by Northern blot hybridization using a 32P-labeled oligonucleotide probe complementary to nucleotides 1–70 of TMV RNA. Positions of size markers are shown on the right with nucleotide length. (Lower) The presence of TMV RNA and the replication proteins in the three samples before MNase treatment. The positions for TMV genomic RNA (G) and the 183-kDa and 126-kDa proteins are indicated. (B) Primer extension analysis. The PMTC fraction and the control SEC fraction were treated with 0, 0.05, 0.1, 0.5, or 1 U/μL MNase. RNA was purified from the reaction mixtures and subjected to primer extension using a 5′-32P-labeled oligonucleotide that was complementary to the nucleotides 61–80 region of TMV RNA. Primer extension products were analyzed by 8 M urea-8% (19:1 bis) PAGE. The band positions are shown on the right with the nucleotide positions that refer to distance from the 5′ terminus. (C) RNase protection analysis. The same set of RNA samples as in B were subjected to RNase protection using a 32P-labeled RNA probe complementary to the nucleotides 31–145 region of TMV RNA. Protected RNA was analyzed by 8 M urea-8% (19:1 bis) PAGE. The band positions are shown on the right with the nucleotide positions that refer to distance from the 5′ terminus. (D) The region of TMV RNA that shows resistance to MNase in the PMTC. Nucleotides marked in green represent the 5′ terminus of the protected fragment revealed by primer extension, and those marked in blue represent the 3′ terminus revealed by RNase protection. The initiation codon for the 126-kDa protein is shown in red. Numbers indicate nucleotide positions with respect to the 5′ terminus. The protected region is roughly indicated by an arrow.