Significance
We show that the genetic susceptibility to the euphoric effects of d-amphetamine also influences the genetic predisposition to schizophrenia and attention deficit hyperactivity disorder (ADHD). These results reinforce the idea that dopamine plays a role in schizophrenia and ADHD; this so-called dopamine hypothesis has been debated for several decades. Specifically, we found that the alleles associated with increased euphoric response to d-amphetamine were associated with decreased risk for schizophrenia and ADHD. These results illustrate how an acute challenge with a pharmacological agent can reveal a genetic predisposition that will manifest itself as psychiatric illness over the lifetime of an individual. Finally, our study offers a relatively novel paradigm for the analysis of endophenotypes for which large sample sizes are not typically available.
Keywords: stimulant, dopamine hypothesis, endophenotype, GWAS, bipolar disorder
Abstract
Here, we extended our findings from a genome-wide association study of the euphoric response to d-amphetamine in healthy human volunteers by identifying enrichment between SNPs associated with response to d-amphetamine and SNPs associated with psychiatric disorders. We found that SNPs nominally associated (P ≤ 0.05 and P ≤ 0.01) with schizophrenia and attention deficit hyperactivity disorder were also nominally associated with d-amphetamine response. Furthermore, we found that the source of this enrichment was an excess of alleles that increased sensitivity to the euphoric effects of d-amphetamine and decreased susceptibility to schizophrenia and attention deficit hyperactivity disorder. In contrast, three negative control phenotypes (height, inflammatory bowel disease, and Parkinson disease) did not show this enrichment. Taken together, our results suggest that alleles identified using an acute challenge with a dopaminergic drug in healthy individuals can be used to identify alleles that confer risk for psychiatric disorders commonly treated with dopaminergic agonists and antagonists. More importantly, our results show the use of the enrichment approach as an alternative to stringent standards for genome-wide significance and suggest a relatively novel approach to the analysis of small cohorts in which intermediate phenotypes have been measured.
Genome-wide association studies (GWAS) implicitly assume that all SNPs in the genome are equally likely to be causal, although most SNPs are unlikely to have any functional consequences. Studies from our groups and others have shown the use of incorporating prior information about SNPs into the genetic analysis of complex traits, including autism and bipolar disorder (1–6). These studies have shown that there is an enrichment of SNPs with functional consequences (e.g., expression quantitative trait loci) among SNPs modestly associated with a broad spectrum of complex traits.
We recently conducted, to our knowledge, the first GWAS of an intermediate pharmacogenetic phenotype, namely the acute subjective response to a drug of abuse, d-amphetamine, in a sample of 381 healthy human volunteers (7). We identified only one genome-wide significant association, and no replication samples were available; thus, the results were difficult to interpret. In the present study, we sought to further interrogate the numerous nominally significant associations from our d-amphetamine response GWAS. We hypothesized that nominally significant associations would be mostly false positives but also, would be enriched for true positives.
Amphetamine produces its subjective and behavioral effects in part by increasing synaptic levels of dopamine (8). We took advantage of prior GWASs for psychiatric disorders to identify a subset of SNPs that showed nominal association with both amphetamine response and psychiatric disorders in which dopaminergic signaling is also hypothesized to play an important role. We predicted that, if these different phenotypes had shared susceptibility alleles, then we would observe more overlapping SNPs than expected by chance. We also predicted that such an enrichment phenomenon would have a consistent direction.
Results
SNPs Associated with the Euphoric Response to d-Amphetamine Are Enriched for SNPs Associated with Protection from Schizophrenia.
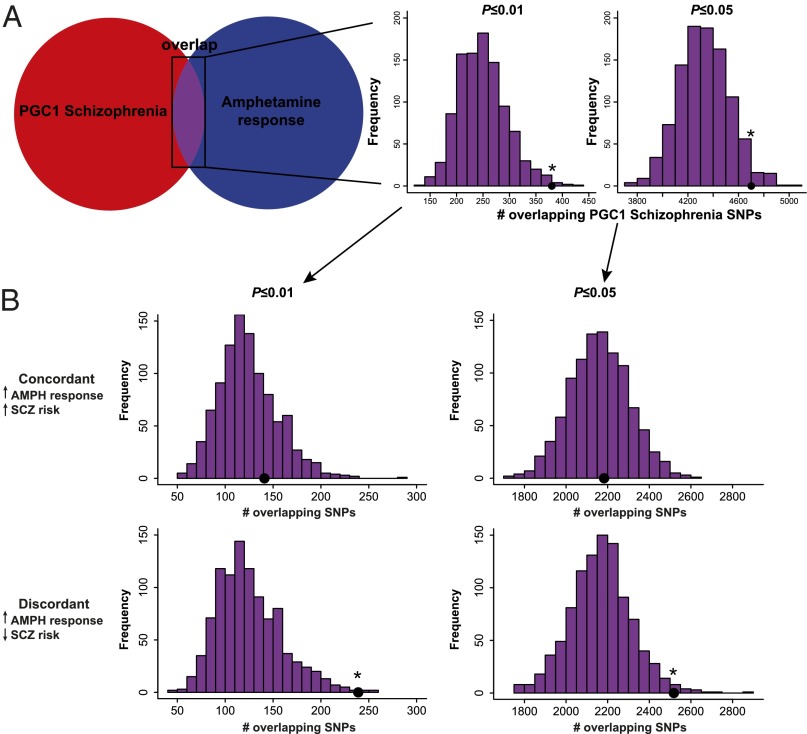
In the enrichment analysis, we observed a statistically significant enrichment of schizophrenia-associated SNPs from the Genetic Association Information Network (GAIN) sample among our associations with amphetamine response at both the P ≤ 0.01 and P ≤ 0.05 thresholds (empirical P = 0.043 and P = 0.005 respectively (Fig. 1). Fig. 2 displays the results from the enrichment analysis of schizophrenia-associated SNPs from Psychiatric Genomics Consortium phase 1 (PGC1), which includes the GAIN dataset as well as a number of additional cohorts. Replicating the results that we initially observed in the GAIN sample, we found a significant enrichment of schizophrenia-associated SNPs among the SNPs associated with amphetamine response at both the P ≤ 0.01 and P ≤ 0.05 thresholds (empirical P = 0.007 and P = 0.033, respectively) (Fig. 2A).
Fig. 1.
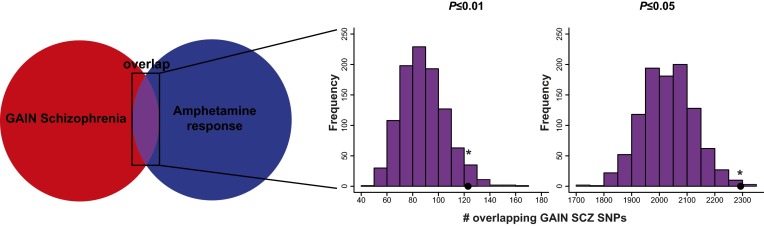
SNPs associated with the euphoric response to d-amphetamine are enriched for SNPs associated with schizophrenia in the GAIN schizophrenia sample. A schematic representation of the enrichment analysis is shown in Left. There was a significant enrichment of SNPs nominally associated with schizophrenia among SNPs nominally associated with the euphoric response to d-amphetamine; the enrichment was significant with P value thresholds of (Center) P ≤ 0.01 and (Right) P ≤ 0.05. The black dots represent the number of overlapping SNPs. The histograms represent the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. SCZ, schizophrenia. *P < 0.05.
Fig. 2.
SNPs associated with the euphoric response to d-amphetamine are enriched among SNPs associated with protection from schizophrenia. A shows a schematic representation of the enrichment analysis. There was a significant enrichment of SNPs that were nominally associated with schizophrenia from the PGC1 Schizophrenia sample among SNPs nominally associated with the euphoric response to d-amphetamine; the enrichment was significant with P value thresholds of (Center) P ≤ 0.01 or (Right) P ≤ 0.05. The black dots represent the observed number of overlapping SNPs. The histograms represent the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. B shows the same analysis as A, except that SNPs were only considered if they were (Upper) concordant in direction or (Lower) discordant in direction. These results indicate that the discordant SNPs are responsible for the enrichment observed in A. AMPH, d-amphetamine; SCZ, schizophrenia. *P < 0.05.
We hypothesized that, if the enrichment phenomena were based on a real biological phenomenon, there would be a consistent relationship between the direction of the effect (positive or negative) of alleles on risk for schizophrenia and sensitivity to the euphoric effects of amphetamine. To test this hypothesis, we performed two analyses: one analysis in which alleles that increased the risk for schizophrenia also increased amphetamine response (concordant) and one analysis in which alleles that increased the risk for schizophrenia decreased amphetamine response (discordant). This analysis could not be performed in the GAIN schizophrenia study, because odds ratios were unavailable. In the PGC1 schizophrenia dataset, we found that 239 of 380 SNPs (62.9%) that constituted the enriched set at the P ≤ 0.01 threshold had discordant direction between the two datasets. Although modest, this enrichment was unambiguously significant compared with the permutation-derived expected distribution of SNPs with discordant direction alleles (empirical P = 0.004) (Fig. 2B). No such enrichment was seen for 141 concordant SNPs (empirical P = 0.269) (Fig. 2B). A similar result was observed when using the P ≤ 0.05 threshold (empirical P = 0.017 for discordant SNPs and empirical P = 0.440 for concordant SNPs) (Fig. 2B). Therefore, the significant enrichment of schizophrenia-associated SNPs among amphetamine-associated SNPs was driven by discordant alleles.
SNPs Associated with the Euphoric Response to d-Amphetamine Are Enriched for SNPs Associated with Protection from Attention Deficit Hyperactivity Disorder.
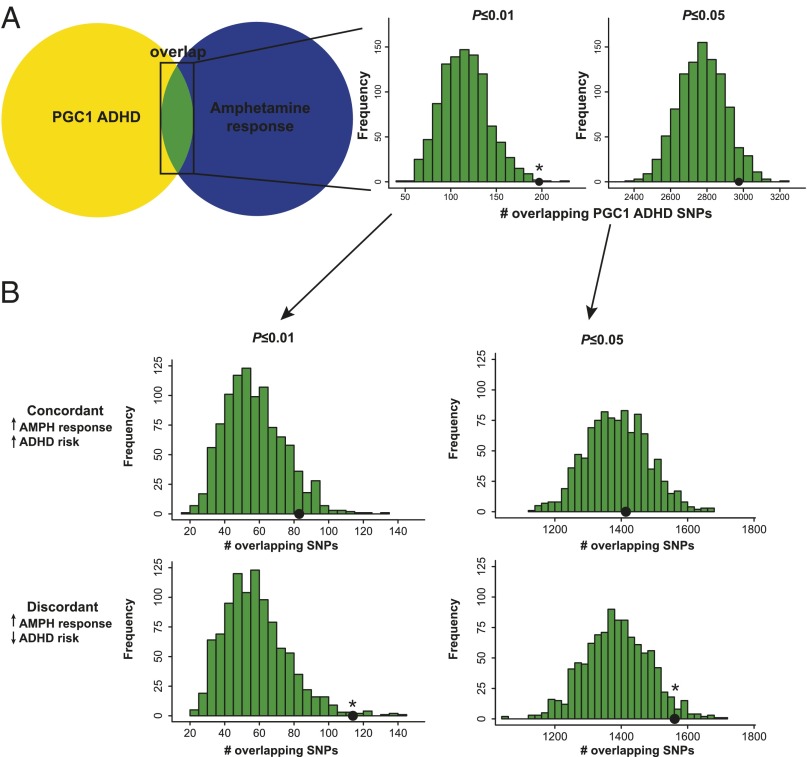
We observed significant enrichment of attention deficit hyperactivity disorder (ADHD)-associated SNPs among the SNPs associated with amphetamine response at both the P ≤ 0.01 and P ≤ 0.05 thresholds (empirical P = 0.011 and P = 0.038, respectively) (Fig. 3A). As with schizophrenia, we hypothesized that there would be a consistent direction of the effects among the overlapping SNPs. Indeed, we found that 114 of 197 overlapping SNPs (57.9%, P ≤ 0.01 threshold) (Fig. 3A) had discordant effects (empirical P = 0.011) (Fig. 3B). No such enrichment was seen for concordant SNPs (empirical P = 0.087) (Fig. 3B). Thus, alleles that decreased risk for ADHD were associated with increased amphetamine response. Similar results were observed at the P ≤ 0.05 threshold (empirical P = 0.038 for discordant SNPs and empirical P = 0.394 for concordant SNPs) (Fig. 3B).
Fig. 3.
SNPs associated with the euphoric response to d-amphetamine are enriched among SNPs associated with protection from ADHD. A shows a schematic representation of the enrichment analysis. There was a significant enrichment of SNPs that were nominally associated with ADHD from the PGC1 ADHD sample among SNPs nominally associated with the euphoric response to d-amphetamine. The results were (Center) significant at the P ≤ 0.01 threshold and (Right) borderline significant at the P ≤ 0.05 threshold. The black dots represent the observed number of overlapping SNPs. The histograms represent the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. B shows the same analysis as A, except that SNPs were only considered if they were (Upper) concordant in direction or (Lower) discordant in direction. These results indicate that the discordant SNPs are responsible for the enrichment observed in A. AMPH, d-amphetamine. *P < 0.05.
SNPs Associated with the Euphoric Response to d-Amphetamine Are Not Enriched for SNPs Associated with Three Negative Control Phenotypes.
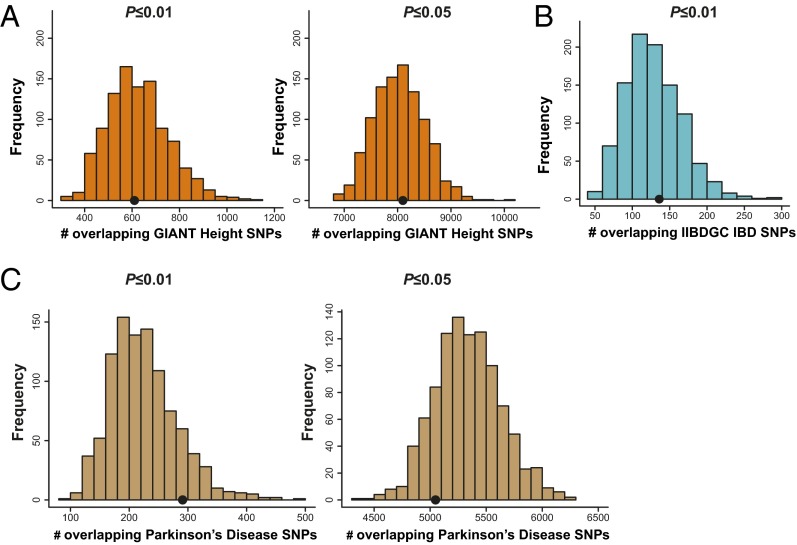
We considered the possibility that enrichment might be caused by linkage disequilibrium (LD) structure or some unexpected artifact not properly accounted for by the permutation analysis and thus, would be observed in any large GWAS. To evaluate this possibility, we examined enrichment in three negative control phenotypes for which large samples were available. We found no significant enrichment of SNPs associated with height at the P ≤ 0.01 or P ≤ 0.05 thresholds (Fig. 4A) (P = 0.518 and P = 0.441, respectively). Similarly, there was no significant enrichment of SNPs associated with inflammatory bowel disease at the P ≤ 0.01 threshold (Fig. 4B) (empirical P = 0.391); data for inflammatory bowel disease at the P ≤ 0.05 threshold were not available. Additionally, we saw no enrichment for Parkinson disease-associated SNPs at either the P ≤ 0.01 or P ≤ 0.05 thresholds (Fig. 4C) (P = 0.126 and P = 0.836, respectively).
Fig. 4.
SNPs associated with the euphoric response to d-amphetamine (P ≤ 0.01 and P ≤ 0.05) do not show enrichment among SNPs associated with height, inflammatory bowel disease, or Parkinson disease. We performed these analyses as a negative control. A shows the results for the height enrichment analysis. Results from the P ≤ 0.01 threshold are shown in Left, and results from the P ≤ 0.05 threshold are shown in Right. The black dots represent the observed count of height-associated SNPs among associations with d-amphetamine response. The histograms represent the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. B shows the results for the inflammatory bowel disease enrichment analysis [P ≤ 0.01 threshold; P ≤ 0.05 results were not available from International Inflammatory Bowel Disease Genetics Consortium (IIBDGC)]. The black dot represents the observed count of inflammatory bowel disease-associated SNPs among associations with d-amphetamine response. The histogram represents the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. None of these results were significant. C shows the results for the Parkinson disease enrichment analysis. Results from the P ≤ 0.01 threshold are shown in Left, and results from the P ≤ 0.05 threshold are shown in Right. The black dots represent the observed count of Parkinson disease-associated SNPs among associations with d-amphetamine response. The histograms represent the null distribution of overlapping SNPs generated from 1,000 random permutations of the amphetamine data. GIANT, Genetic Investigation of Anthropometric Traits.
In terms of directionality in the negative control samples, we found no significant enrichment of concordant or discordant SNPs in the Parkinson disease dataset. We were unable to obtain directional information for the height and inflammatory bowel disease datasets. However, we were able to obtain directional information for a Crohn disease GWAS dataset that largely overlaps with a subset of the inflammatory bowel disease sample (9). Using that dataset, we observed no significant overall enrichment and no significant enrichment of concordant or discordant SNPs.
Similar Results Are Observed When Imputed SNPs from the Amphetamine Response Dataset Are Excluded.
All results presented were derived from analyses using amphetamine response data that consist of a mixture of directly genotyped and imputed SNPs. To assess the possibility that an artifact related to imputation had caused the observed enrichment, we conducted similar analyses that were restricted to directly genotyped SNPs in the amphetamine response dataset; these results were not meaningfully different (Fig. S1). Thus, these results do not seem to be an artifact of imputation.
Enrichment of Schizophrenia and ADHD-Associated SNPs Is Observed in Replication Samples.
To replicate our findings of enrichment for schizophrenia associated SNPs in the GAIN and PGC1 datasets, we obtained an additional replication dataset [Swedish schizophrenia sample (10)] and repeated our analyses in the replication sample alone and the combined meta-analysis sample (PGC1 schizophrenia + Swedish schizophrenia). When considering only the Swedish schizophrenia sample, we observed borderline significant enrichment at the P ≤ 0.05 threshold (P = 0.067); when we performed the same analysis in the meta-analysis sample (PGC1 schizophrenia + Swedish schizophrenia), we found that the strength of enrichment improved (P = 0.021) compared with the same analysis in the PGC1 schizophrenia sample alone. We also found that the strength of enrichment among the discordant SNPs was slightly improved in this larger meta-analysis sample (P = 0.016) compared with the results from the PCG1 schizophrenia data.
Similarly, we were able to replicate our findings in a newer ADHD replication dataset [Psychiatric Genomics Consortium phase 2 (PGC2) ADHD] using the P ≤ 0.05 threshold. In this case, we did not observe a significant enrichment when using only the ADHD replication dataset (PGC2 ADHD); however, we did observe a nearly significant enrichment of discordant direction SNPs (P = 0.060). Similarly, in the meta-analysis sample (PGC1 ADHD + PGC2 ADHD), we observed an even more significant enrichment of discordant direction SNPs (P = 0.010) in the meta-analysis sample compared with the PCG1 ADHD sample alone.
SNPs Associated with the Increased Euphoric Response to d-Amphetamine Are Enriched for SNPs That Confer Protection from Bipolar Disorder.
We hypothesized that SNPs associated with the euphoric response to amphetamine may also be enriched for SNPs associated with bipolar disorder. We did not observe an overall significant enrichment (Fig. S2). However, when we stratified SNPs by concordant vs. discordant, we again observed a significant enrichment of discordant SNPs at both the P ≤ 0.01 and P ≤ 0.05 thresholds (empirical P = 0.018 and P = 0.045, respectively) (Fig. S2).
A Subset of the SNPs That Are Associated with the Euphoric Response to d-Amphetamine Are Enriched for SNPs That Confer Protection from Both Schizophrenia and ADHD.
We were interested in testing whether any of the SNPs that overlapped with d-amphetamine response were shared with both schizophrenia and ADHD. Shared SNPs would suggest shared biology, potentially related to dopaminergic function. We found suggestive evidence for enrichment of SNPs shared among all three phenotypes (amphetamine response, schizophrenia, and ADHD; P = 0.062) (Fig. S3). When we tested only concordant SNPs (increased amphetamine response and increased risk for both schizophrenia and ADHD) and discordant SNPs (increased amphetamine response and decreased risk for both schizophrenia and ADHD), we only found significant enrichment for discordant SNPs (P = 0.029) (Fig. S3), similar to results for schizophrenia and ADHD. This analysis identified a small subset of SNPs that is likely to contribute to enhanced euphoric responses to d-amphetamine and decreased risk for schizophrenia and ADHD. This result is interesting in light of the results from the PGC Cross-Disorder group, which showed no genetic overlap between schizophrenia and ADHD (6).
Discussion
Our results show that SNPs associated with response to a dopaminergic drug challenge (d-amphetamine) are enriched for SNPs associated with psychiatric disorders that are treated with dopamine agonists (ADHD) and antagonists (schizophrenia). Rather than identifying a few SNPs with a high degree of statistical confidence, our method is intended to identify a heterogeneous collection of SNPs that is made up of both true- and false-positive associations. We show that this enrichment was caused by alleles that increased the euphoric response to amphetamine and decreased the risk for both schizophrenia and ADHD. In contrast, no enrichment was observed for concordant SNPs or any nonpsychiatric phenotypes. We also showed that the results were not an artifact of imputation and that these effects could be replicated in multiple samples.
Of the theories regarding the underlying mechanisms for schizophrenia, the so-called dopamine hypothesis has been the most enduring (11, 12). Although this theory is still under debate (13, 14), several lines of evidence lend credence to the hypothesis. For example, the efficacy of typical antipsychotic drugs is almost linearly related to their affinity for the dopamine D2 receptor (15). Additionally, when high doses of amphetamine are ingested for a protracted period, psychotic symptoms can develop (16). Several studies have shown increased striatal dopamine release in response to a d-amphetamine challenge in schizophrenics and consequently, a worsening of symptoms (17, 18). Our study adds genetic evidence to support the dopaminergic hypothesis of schizophrenia using a cohort of healthy volunteers carefully screened against Axis I psychiatric disorders.
A dopamine hypothesis of ADHD has also been proposed and challenged (19, 20). ADHD is often treated with methylphenidate or amphetamine products (d-amphetamine, mixed amphetamine salts, or lisdexamfetamine) (21). The therapeutic effects of these drugs are believed to be caused by their ability to increase the synaptic availability of dopamine. Interestingly, our results suggest that insensitivity to a drug that is used to treat ADHD might be a genetic risk factor for ADHD; however, it is important to note that we examined sensitivity to the euphoric effects of amphetamine and not sensitivity to its therapeutic effects. Our results are consistent with studies that have shown a protective effect from substance use disorders in stimulant-treated adolescents with ADHD (22, 23).
A puzzling feature of our results is that we saw enrichment of protective alleles for both schizophrenia and ADHD among our top associations with acute amphetamine response, whereas a simplistic understanding of these disorders suggests different types of dopamine dysregulation: excess dopamine in schizophrenia vs. dopamine deficit in ADHD. There is mixed evidence for shared genetic risk for schizophrenia and ADHD. A higher incidence of ADHD symptoms has been observed among relatives of schizophrenic patients compared with healthy controls (24) as well as increased risk for schizophrenia among relatives of individuals with ADHD (25). A recent polygenic risk score analysis identified shared genetic susceptibility between schizophrenia and ADHD (26). However, another recent study did not identify significant polygenic risk overlap for schizophrenia and ADHD (6), and a different recent study found no significant genetic correlation estimated from SNP heritabilities for the two disorders (27). Our approach is different, because we are examining only the subset of SNPs that is associated with both amphetamine response and these psychiatric disorders, which may explain the discrepancy between our results and these two recent studies and may identify another advantage of our approach.
These data suggest that our acute amphetamine response phenotype may be viewed as an endophenotype for schizophrenia and ADHD. Whereas prior definitions of endophenotypes have focused on cosegregation of the putative endophenotype and the disease phenotype, we examined associations at SNPs throughout the genome to establish a genetic link between amphetamine response with both schizophrenia and ADHD. Our sample was specifically screened to exclude individuals with Axis I disorders, which should have depleted the number of risk alleles present in this population. The results suggest a relatively novel approach to the empirical validation of endophenotypes.
Comorbidity of ADHD and bipolar disorder has been reported in the literature (28), and thus, we considered the possibility of enrichment of bipolar disorder-associated SNPs and amphetamine response-associated SNPs. Although we did not observe overall enrichment, we did observe directionality, with significant enrichment of discordant SNPs at two P value thresholds. These results suggest that, in addition to schizophrenia and ADHD, the acute amphetamine response phenotype may also be an endophenotype for bipolar disorder (29).
We initially conceived of the acute response to amphetamine as an intermediate phenotype for drug abuse. However, our results suggest that acute drug challenge phenotypes may be useful in identifying SNPs that are functionally relevant to psychiatric disorders. Based on this study, it may be reasonable to ask whether sensitivity to therapeutic drugs (or drugs that cause worsening of symptoms) may uncover alleles that confer risk or protection for other disorders. Whether acute amphetamine response is indeed a useful intermediate phenotype for drug abuse or other disorders may be determined in future studies; related research examining the euphoric response to alcohol has proven fruitful (30–34).
Our amphetamine response GWAS was based on a relatively small sample. Lack of power is likely to contribute to the inability to achieve signals that survive multiple testing corrections in the GWAS of psychiatric phenotypes (35). By taking an enrichment approach, we were able to capitalize on associations that did not meet stringent genome-wide significance criteria but were nominally associated with amphetamine response. Our results suggest that the enrichment approach is complementary to the traditional GWAS approach and a valuable secondary analysis. In contrast to GWAS, which aims to identify specific SNPs, the power of our method is that it can draw biological inferences from a heterogeneous set of SNPs composed of both true and false positives. However, this method is unable to distinguish between these two categories.
Although our study is not without limitations, we considered several alternative explanations for our observations, but none proved credible. One possibility was that results from any two GWAS may overlap because of LD patterns. By using permutation, we preserved the LD structure among the SNPs being tested, which should guard against such a phenomenon. This possibility is further addressed by the directional analyses and our use of negative control phenotypes. We considered the possibility that the enrichment that we observed was driven by functional brain SNPs (e.g., expression quantitative trait loci) that would be enriched for any brain disease. However, we saw no enrichment for Parkinson disease-associated SNPs, suggesting that our results are specific to schizophrenia and ADHD; the results from our directional analyses of schizophrenia and ADHD further dispute the possibility that the overlapping SNPs are important for all brain diseases. We were also concerned that artifacts caused by imputation could bias our results. However, we observed similar results when we considered only SNPs that were directly genotyped in the amphetamine response sample; permutation should further guard against any such artifacts (Fig. S1). Our results are further strengthened by the fact that they were observed in multiple datasets.
By examining our GWAS results through the lens of enrichment, we were able to interrogate results that do not meet stringent criteria for statistical significance. Our results suggest that alleles identified using an acute drug challenge can be used to identify alleles that influence risk for psychiatric disorders. Our results also support the dopamine hypotheses of schizophrenia and ADHD. Ultimately, this study shows that additional secondary analyses of GWAS results may provide new insights into the biology of psychiatric disorders. These results also suggest a useful and generalizable method for the genetic analysis of modestly sized intermediate phenotypes that are unlikely to yield genome-wide significant results and for which replication samples are not typically available.
Materials and Methods
Genetics of Amphetamine Dataset.
Study details are provided in the work by Hart et al. (7). This study was approved by the Institutional Review Board of The University of Chicago and was carried out in accordance with the Helsinki Declaration of 1975. Briefly, 381 healthy volunteers attended three separate 4-h sessions, during which they received d-amphetamine (placebo, 10 mg, or 20 mg) under double blind conditions and subjective self-report questionnaires at regular intervals: the Profile of Mood States (36), Drug Effects Questionnaire (37), and Addiction Research Center Inventory (38). Sparse factor analysis (39) was used to reduce the dimensionality of the phenotype data to a small number of factors that explained both drug response and baseline characteristics of the sample. For the present study, we limited our analyses to the 10-mg [d-amphetamine] response factor. This factor, hereafter referred to as amphetamine response, was one of the most interpretable factors, reflecting the subjective euphoric response to amphetamine, and it showed the strongest association signal (7). Subjects were genotyped using Affymetrix 6.0 arrays. Imputation was performed using the HapMap3 and 1000 Genomes reference panels (40, 41). Self-reported ancestry was confirmed by analysis with the SMARTPCA component of EIGENSOFT (42). The sample used in the current study was restricted to participants of European ancestry (n = 325). After quality control and imputation, 5,974,669 SNPs were available for analysis. The samples used for the enrichment analysis are shown in Table S1; additional details are given in the SI Materials and Methods.
Data Preparation.
In the Genetics of Amphetamine dataset, SNPs with minor allele frequencies < 0.01 were removed. Genotypes were converted into PLINK format with GTOOL (www.well.ox.ac.uk/∼cfreeman/software/gwas/gtool.html) with a threshold of 0.8 specified; markers with missing rates > 10% were excluded. The amphetamine response phenotype was permuted 1,000 times using the “make-perm-pheno” command in PLINK (43), and association testing was run with each of these 1,000 permuted phenotypes with the PLINK “assoc” command. The numbers of SNPs available for the enrichment analysis are listed in Table S2.
Enrichment Analysis.
The number of SNPs that overlapped between the amphetamine response results and the results for each of the phenotypes described above was recorded (for both the P ≤ 0.01 and P ≤ 0.05 thresholds). Next, the number of overlapping SNPs in each permuted dataset (n = 1,000) was recorded, yielding the expected null distribution. The empirical P value was computed as the fraction of permutations where the number of overlapping SNPs matched or exceeded the observed count. A statistically significant enrichment was defined as an enrichment P value < 0.05 (i.e., less than 50 permutations were found with a greater number of overlapping SNPs).
Directionality Analysis.
For the SNPs that overlapped between the phenotypes examined in the enrichment analyses described above, we examined the direction of the effect in both the amphetamine response and the second phenotype. The signs of the logistic regression β-coefficients [i.e., ln(odd ratio)] were used to denote directionality. The Z scores from the PGC1 ADHD results were used to denote directionality of the association, with Z score > 0 corresponding to odds ratio > 1. The signs of the β-coefficients or Z scores were flipped if the PGC reference allele did not match the reference allele in the amphetamine response dataset. We recorded the number of concordant SNPs (positive in both samples or negative in both samples) and the number of discordant SNPs (positive in one sample and negative in the other sample) in the real and permuted datasets. This procedure generated the expected null distribution of concordant alleles (e.g., alleles associated with risk as well as heightened response to amphetamine) and the expected null distribution of discordant alleles. Excluding strand ambiguous SNPs had no effect on our results. The empirical P value was computed as the proportion of permutations where the number of overlapping SNPs matched or exceeded the count observed in the real data.
Replication Analyses.
Enrichment and directionality analyses were performed as described above in the replication samples alone (Swedish schizophrenia study and PGC2 ADHD) and the combined meta-analysis samples (PGC1 schizophrenia + Swedish schizophrenia and PGC1 ADHD + PGC2 ADHD). Meta-analysis was performed with the “meta-analysis” command in PLINK.
Supplementary Material
Acknowledgments
We thank Dr. Peter Visscher and Dr. Naomi Wray for helpful discussions. This work was supported by National Institutes of Health Grants P50MH094267 (to N.J.C., E.R.G., A.A.P., and A.B.H.); DA007255 (to A.B.H.); GM61393, MH094267, MH101820, and MH090937 (all to E.R.G. and N.J.C.); HG006265 (to B.E.E.); MH062873 and MH059126 (both to S.V.F.); DA02812 and DA032015 (both to H.d.W.); and DA021336 and DA027545 (both to A.A.P.). Funding for the Swedish study was provided by National Institute of Mental Health Grants R01 MH095034 (to P.S.) and R01 MH077139 (to P.F.S.); the Stanley Center for Psychiatric Research, the Sylvan Herman Foundation, the Karolinska Institutet, Karolinska University Hospital, the Swedish Research Council, the Swedish County Council, and the Söderström Königska Foundation (all C.M.H.); and Netherlands Organization for Scientific Research Grant 645-000-003. B.E.E. was also funded through the Bioinformatics Research Development Fund supported by Kathryn and George Gould.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
1A complete list of the Psychiatric Genomics Consortium: ADHD Subgroup members can be found in the SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318810111/-/DCSupplemental.
References
- 1.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6(4):e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis LK, et al. Loci nominally associated with autism from genome-wide analysis show enrichment of brain expression quantitative trait loci but not lymphoblastoid cell line expression quantitative trait loci. Mol Autism. 2012;3(1):3. doi: 10.1186/2040-2392-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gamazon ER, Huang RS, Cox NJ, Dolan ME. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc Natl Acad Sci USA. 2010;107(20):9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamazon ER, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18(3):340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86(4):581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smoller JW, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium Genetic Risk Outcome of Psychosis (GROUP) Consortium Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart AB, et al. Genome-wide association study of d-amphetamine response in healthy volunteers identifies putative associations, including cadherin 13 (CDH13) PLoS ONE. 2012;7(8):e42646. doi: 10.1371/journal.pone.0042646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuczenski R, Segal DS. Effects of methylphenidate on extracellular dopamine, serotonin, and norepinephrine: Comparison with amphetamine. J Neurochem. 1997;68(5):2032–2037. doi: 10.1046/j.1471-4159.1997.68052032.x. [DOI] [PubMed] [Google Scholar]
- 9.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42(12):1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ripke S, et al. Multicenter Genetic Studies of Schizophrenia Consortium Psychosis Endophenotypes International Consortium Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson A, Lindqvist M. Effect of chlorpromazine or haloperidol on formation of 3‐methoxytyramine and normetanephrine in mouse brain. Acta Pharmacol Toxicol (Copenh) 1963;20(1963):140–144. doi: 10.1111/j.1600-0773.1963.tb01730.x. [DOI] [PubMed] [Google Scholar]
- 12.Van Rossum JM. The significance of dopamine-receptor blockade for the mechanism of action of neuroleptic drugs. Arch Int Pharmacodyn Ther. 1966;160(2):492–444. [PubMed] [Google Scholar]
- 13.Abi-Dargham A. Do we still believe in the dopamine hypothesis? New data bring new evidence. Int J Neuropsychopharmacol. 2004;7(Suppl 1):S1–S5. doi: 10.1017/S1461145704004110. [DOI] [PubMed] [Google Scholar]
- 14.Kendler KS, Schaffner KF. The dopamine hypothesis of schizophrenia: An historical and philosophical analysis. Philos Psychiatry Psychol. 2011;18(1):41–63. [Google Scholar]
- 15.Peroutka SJ, Synder SH. Relationship of neuroleptic drug effects at brain dopamine, serotonin, alpha-adrenergic, and histamine receptors to clinical potency. Am J Psychiatry. 1980;137(12):1518–1522. doi: 10.1176/ajp.137.12.1518. [DOI] [PubMed] [Google Scholar]
- 16.Snyder SH. Amphetamine psychosis: A “model” schizophrenia mediated by catecholamines. Am J Psychiatry. 1973;130(1):61–67. doi: 10.1176/ajp.130.1.61. [DOI] [PubMed] [Google Scholar]
- 17.Abi-Dargham A, et al. Increased striatal dopamine transmission in schizophrenia: Confirmation in a second cohort. Am J Psychiatry. 1998;155(6):761–767. doi: 10.1176/ajp.155.6.761. [DOI] [PubMed] [Google Scholar]
- 18.Laruelle M, et al. Single photon emission computerized tomography imaging of amphetamine-induced dopamine release in drug-free schizophrenic subjects. Proc Natl Acad Sci USA. 1996;93(17):9235–9240. doi: 10.1073/pnas.93.17.9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonon F. The dopaminergic hypothesis of attention-deficit/hyperactivity disorder needs re-examining. Trends Neurosci. 2009;32(1):2–8. doi: 10.1016/j.tins.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Wender PH. Minimal Brain Dysfunction in Children. New York: Wiley Interscience; 1971. [Google Scholar]
- 21.Warikoo N, Faraone SV. Background, clinical features and treatment of attention deficit hyperactivity disorder in children. Expert Opin Pharmacother. 2013;14(14):1885–1906. doi: 10.1517/14656566.2013.818977. [DOI] [PubMed] [Google Scholar]
- 22.Biederman J. Pharmacotherapy for attention-deficit/hyperactivity disorder (ADHD) decreases the risk for substance abuse: Findings from a longitudinal follow-up of youths with and without ADHD. J Clin Psychiatry. 2003;64(Suppl 11):3–8. [PubMed] [Google Scholar]
- 23.Groenman AP, et al. Stimulant treatment for attention-deficit hyperactivity disorder and risk of developing substance use disorder. Br J Psychiatry. 2013;203(2):112–119. doi: 10.1192/bjp.bp.112.124784. [DOI] [PubMed] [Google Scholar]
- 24.Keshavan MS, Sujata M, Mehra A, Montrose DM, Sweeney JA. Psychosis proneness and ADHD in young relatives of schizophrenia patients. Schizophr Res. 2003;59(1):85–92. doi: 10.1016/s0920-9964(01)00400-5. [DOI] [PubMed] [Google Scholar]
- 25.Larsson H, et al. Risk of bipolar disorder and schizophrenia in relatives of people with attention-deficit hyperactivity disorder. Br J Psychiatry. 2013;203(2):103–106. doi: 10.1192/bjp.bp.112.120808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamshere ML, et al. Shared polygenic contribution between childhood attention-deficit hyperactivity disorder and adult schizophrenia. Br J Psychiatry. 2013;203(2):107–111. doi: 10.1192/bjp.bp.112.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faraone SV, Biederman J, Wozniak J. Examining the comorbidity between attention deficit hyperactivity disorder and bipolar I disorder: A meta-analysis of family genetic studies. Am J Psychiatry. 2012;169(12):1256–1266. doi: 10.1176/appi.ajp.2012.12010087. [DOI] [PubMed] [Google Scholar]
- 29.Kelsoe JR, Niculescu AB., 3rd Finding genes for bipolar disorder in the functional genomics era: From convergent functional genomics to phenomics and back. CNS Spectr. 2002;7(3):215–216, 223–226. doi: 10.1017/s1092852900017582. [DOI] [PubMed] [Google Scholar]
- 30.Ray LA, Hutchison KE. A polymorphism of the mu-opioid receptor gene (OPRM1) and sensitivity to the effects of alcohol in humans. Alcohol Clin Exp Res. 2004;28(12):1789–1795. doi: 10.1097/01.alc.0000148114.34000.b9. [DOI] [PubMed] [Google Scholar]
- 31.Ramchandani VA, et al. A genetic determinant of the striatal dopamine response to alcohol in men. Mol Psychiatry. 2011;16(8):809–817. doi: 10.1038/mp.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setiawan E, et al. The effect of naltrexone on alcohol’s stimulant properties and self-administration behavior in social drinkers: Influence of gender and genotype. Alcohol Clin Exp Res. 2011;35(6):1134–1141. doi: 10.1111/j.1530-0277.2011.01446.x. [DOI] [PubMed] [Google Scholar]
- 33.King AC, de Wit H, McNamara PJ, Cao D. Rewarding, stimulant, and sedative alcohol responses and relationship to future binge drinking. Arch Gen Psychiatry. 2011;68(4):389–399. doi: 10.1001/archgenpsychiatry.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit H, Phillips TJ. Do initial responses to drugs predict future use or abuse? Neurosci Biobehav Rev. 2012;36(6):1565–1576. doi: 10.1016/j.neubiorev.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cichon S, et al. Psychiatric GWAS Consortium Coordinating Committee Genomewide association studies: History, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johanson CE, Uhlenhuth EH. Drug preference and mood in humans: Diazepam. Psychopharmacology (Berl) 1980;71(3):269–273. doi: 10.1007/BF00433061. [DOI] [PubMed] [Google Scholar]
- 37.Chait LD, Fischman MW, Schuster CR. ‘Hangover’ effects the morning after marijuana smoking. Drug Alcohol Depend. 1985;15(3):229–238. doi: 10.1016/0376-8716(85)90002-x. [DOI] [PubMed] [Google Scholar]
- 38.Martin WR, Sloan JW, Sapira JD, Jasinski DR. Physiologic, subjective, and behavioral effects of amphetamine, methamphetamine, ephedrine, phenmetrazine, and methylphenidate in man. Clin Pharmacol Ther. 1971;12(2):245–258. doi: 10.1002/cpt1971122part1245. [DOI] [PubMed] [Google Scholar]
- 39.Engelhardt BE, Stephens M. Analysis of population structure: A unifying framework and novel methods based on sparse factor analysis. PLoS Genet. 2010;6(9):e1001117. doi: 10.1371/journal.pgen.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.