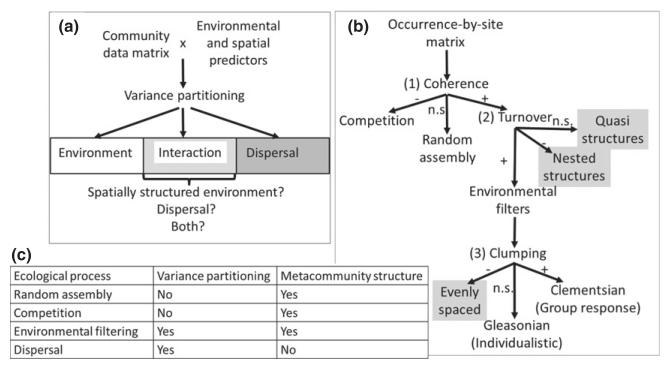
Figure 1. The two metacommunity analyses used in this study of grassland plant communities within the French Alps.
(a) Variance partitioning allows the identification of relevant environmental predictors structuring species composition and separation of the effects of environmental filtering from that of dispersal. However, the interaction term between spatial structure and environmental predictors as well as the unexplained variance in the models give ambiguous results with respect to the four metacommunity processes (dispersal, environmental filtering, biological interactions, and stochastic colonization–extinction dynamics). (b) Analysis of metacommunity structure through coherence, range turnover and boundary clumping using the site-by-species incidence matrix allows some observed patterns to be linked to their possible causal processes: random assembly (non-significant coherence), competitive exclusion (negative coherence) and environmental filtering (Gleasonian or Clementsian responses to environmental gradients). However, some other results cannot be clearly linked to the four metacommunity processes (grey boxes in the figure), and it may be possible that competition and environmental filtering may explain some of them as well. (c) Summary of the processes that can be distinguished by using one or the other type of analysis, showing that their combination opens new avenues in metacommunity analyses. n.s., non-significant.