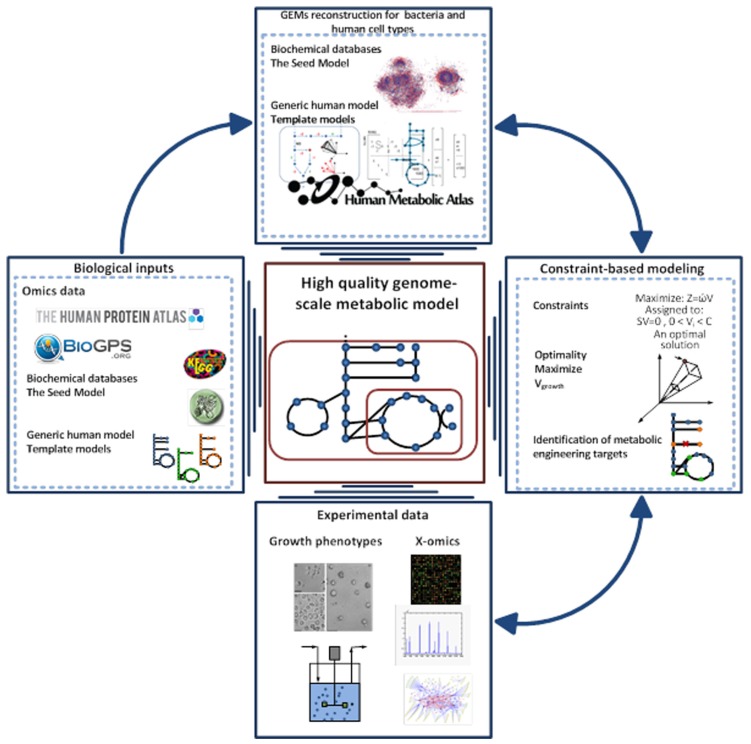
FIGURE 2.
Pipeline for high-quality GEM reconstruction. This approach has been applied to the reconstruction of GEMs for microbes, tissue, and cell types. The Raven Toolbox is mainly applied for GEMs reconstruction based on template models and different databases 35. After having the draft model, sets of quality control can be applied to generate a fully functional model. This toolbox enables integration of omics data for more comprehensive analysis. It can generate the models in different formats, visualizing the simulations and overlaying the model in a metabolic map. Another powerful plug in is the INIT algorithm, which can be used for reconstruction of tissue-specific models based on a human generic metabolic model (Agren et al., 2012). The GEMs for tissues and microbes can be validated through constraint-based modeling approaches and available experimental data.