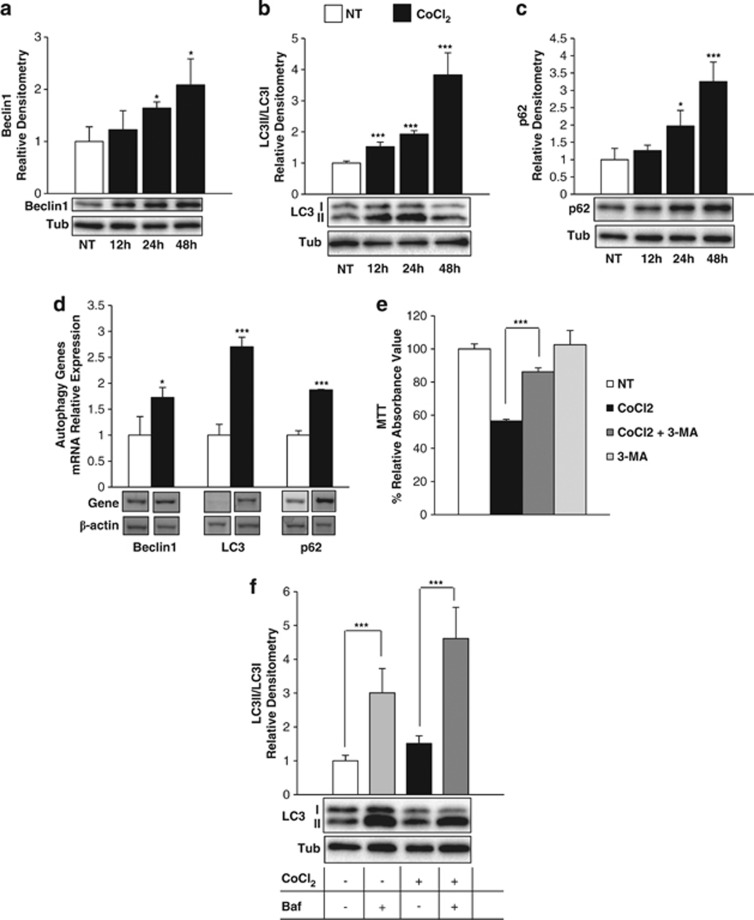
Figure 5.
CoCl2 induces autophagy in H9c2 cardiomyoblasts. Cells were NT (white) or treated with CoCl2 (300 μM, black) for different lengths of time. Protein (a–c) and mRNAs (d) densitometric quantification of autophagic markers: Beclin-1, LC3 and p62. The densitometric quantification of protein LC3II/LC3I ratio is reported. mRNA expression analysis was performed at 48 h. The t-test was performed on treated versus NT cells. (e) Cell viability was measured by MTT. Cells were NT (white) or treated with CoCl2 for 48 h (black), CoCl2+1 mM 3-MA inhibitor of autophagy in the last 12 h (dark grey) and 3-MA alone (light grey). Results are expressed as percentage of MTT reduction, relative to NT control. t-test was calculated comparing CoCl2 versus CoCl2+3-MA. (f) Densitometric quantification of protein LC3II/LC3I ratio in cells cultured in the presence or absence of Baf (50 nM). Cells were treated with CoCl2 for 48 h and Baf was added in the last 6 h. In the graph four conditions of treatment are reported: NT (white), Baf (light grey), CoCl2 (black) and CoCl2+Baf (dark grey). t-test was calculated between NT versus Baf and CoCl2 versus CoCl2+Baf. Tubulin and β-actin were used as loading control for protein and mRNA, respectively. Representative images are reported below each graph. The values are the mean±S.D. of three independent experiments. In all the experiments, except for (e), values are expressed as fold relative to NT. *P<0.05 and ***P<0.005