Figure 3.
The d14-seto Protein.
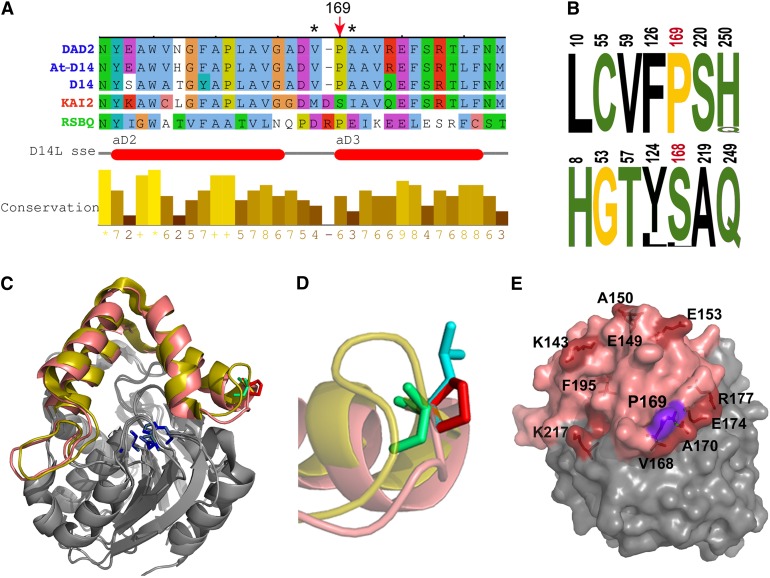
(A) Sequence alignment of the Arabidopsis D14 segment comprising the Pro169Leu mutation, with ortholog sequences petunia DAD2 (Hamiaux et al., 2012), rice D14 (Arite et al., 2009; Gao et al., 2009; Liu et al., 2009; Hamiaux et al., 2012), paralog KAI2 (Waters et al., 2012b), and related bacterial protein RsbQ (Brody et al., 2001). Red arrow indicates Pro-169 and corresponding amino acid Ser-168 in KAI2. Asterisks indicate residues Met-166 and Ile-169, which undergo conformational changes in KAI2 after KAR1 binding (Guo et al., 2013b). Horizontal red bars indicate the position of two of the KAI2 cap α-helices (Kagiyama et al., 2013).
(B) Logos of SDPs that differ in D14 (top) and KAI2 (bottom) ortholog sequences. Numbering corresponds to D14 and KAI2 protein sequences. Pro-169 and Ser-168 are shown in red. Hydrophobic residues are indicated in black, polar residues in green, and Gly and Pro in yellow. Letter size represents percentage of conservation within protein classes.
(C) Front view of D14 (PDB:4ih4) and KAI2 (PDB:3w06) structural alignment. Helical caps of D14 and KAI2 are highlighted in salmon pink and yellow, respectively. Active site residues are in blue, D14 Pro-169 is in red, and KAI2 S168 is in green.
(D) Close-up view and side chain superposition of wild-type D14 P169 (red), mutant Leu-169 (blue), and KAI2 Ser-168 (green). Note that the Pro-169 side chain is exposed to the solvent and that KAI2 loop (yellow) is longer than that of D14.
(E) D14 structure in surface representation. Residues corresponding to residues in KAI2 that undergo side-chain movement after KAR1 binding are labeled and highlighted in red, Pro-169 in purple and cap domain in pink.