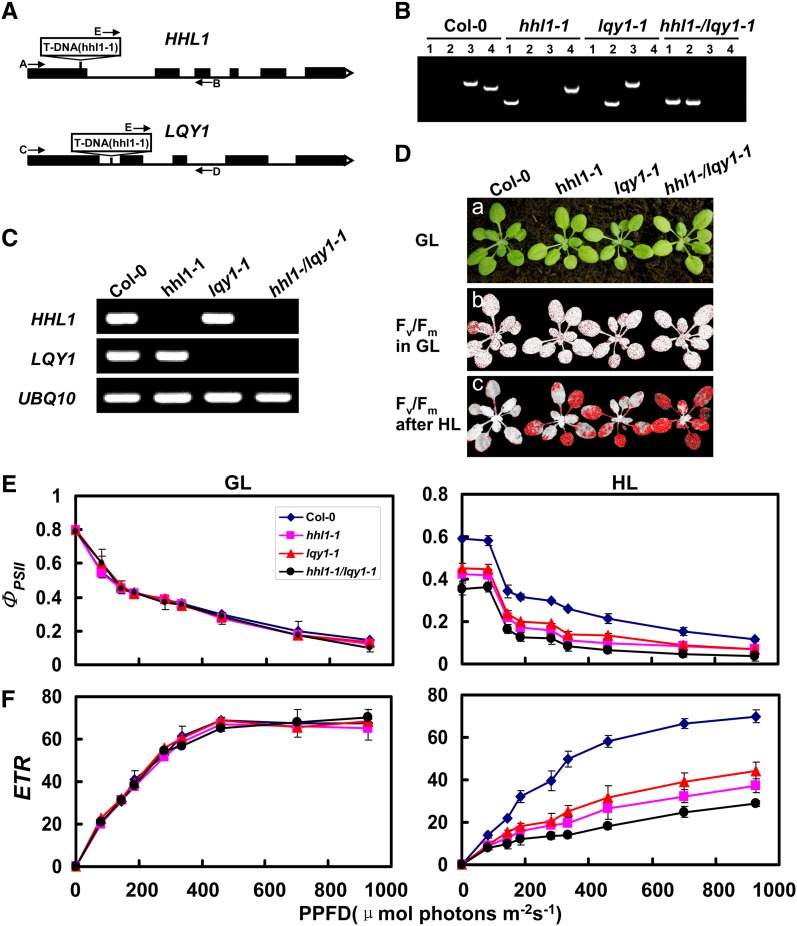
Figure 10.
Phenotypic Analyses of hhl1, lqy1, and hhl1 lqy1 Mutants and Wild-Type Plants.
(A) Schematic diagram of the HHL1 and LQY1 genes. Exons (black boxes) and introns (lines) are indicated. The positions of the T-DNA insertions corresponding to hhl1-1 and lqy1-1 are shown. Arrows indicate the location of primers used for PCR analyses.
(B) PCR analysis of genomic DNA from the wild type and hhl1, lqy1, and hhl1 lqy1 plants confirm the homozygosity of the mutants. Lane 1, amplification with primers E and B; lane 2, amplification with primers E and D; lane 3, amplification with primers A and B; lane 4, amplification with primers C and D.
(C) RT-PCR analysis of HHL1 and LQY1 gene expression, performed with the specific primers for HHL1, LQY1, and UBQ10 genes.
(D) Images in (a) are of 3-week-old wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants under a growth light (GL) conditions. (b) False-color images representing Fv/Fm under growth light conditions in 3-week-old wild-type, hhl1-1, lqy1-1, and hhl1 lqy1 plants. Red pixels indicate that Fv/Fm is below the cutoff value (0.753). (c) False-color images representing Fv/Fm after a 3-h high-light (HL) treatment in 3-week-old wild-type, hhl1-1, lqy1-1, and hhl1 lqy1 plants. Red pixels indicate that Fv/Fm is below the cutoff value (0.439).
(E) and (F) Light-response curves of PSII quantum yield (E) and ETR (F) in wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants in growth light (GL) and after 3-h high-light (HL) treatment. Data for wild-type (Col-0), hhl1-1, lqy1-1, and hhl1 lqy1 plants are presented as mean ± se. Four biological replicates were performed, and similar results were obtained.