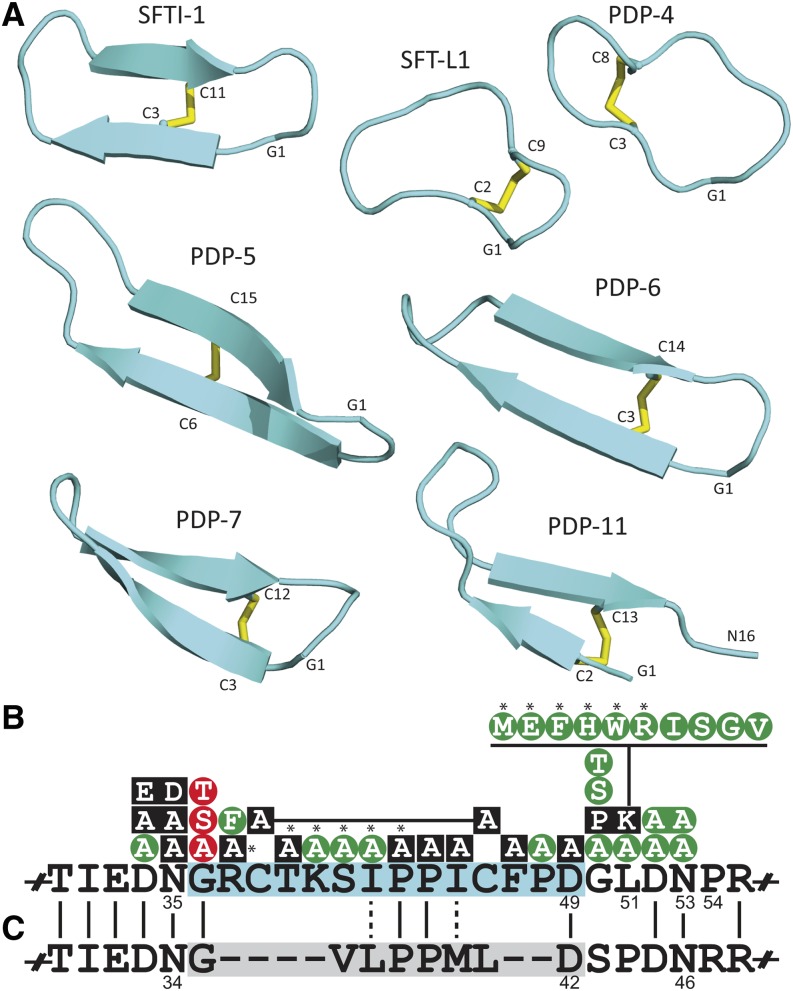
Figure 3.
Structures of PDPs and in Vivo Peptide-Processing Constraints for SFTI-1.
(A) Ribbon representation of NMR-derived structures for novel PDPs alongside sunflower SFTI-1 (Korsinczky et al., 2001) and SFT-L1 (Mylne et al., 2011).
(B) In vivo mutagenesis data from previous work (Mylne et al., 2011) and this study (asterisked) reveals which changes are tolerated biologically (green), perturb peptide processing (black), or specifically perturb the cyclization reaction (red).
(C) PawL1 from A. montana shares many of the critical residues for peptide maturation but lacks Cys residues and does not make a stable peptide.