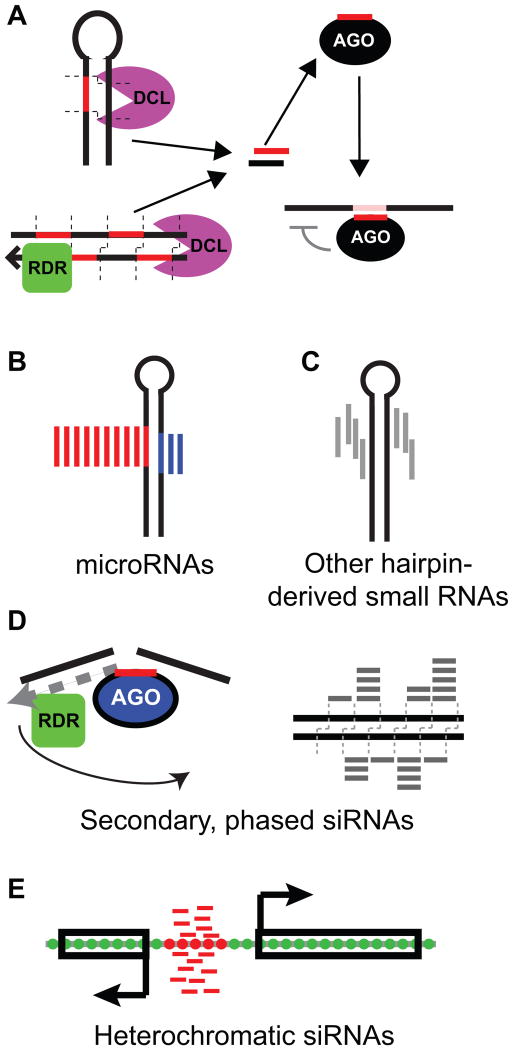
Figure 1.
Overview of plant small RNAs.
A. Processing and usage of plant small RNAs. Substrates are either the stems of hairpins (top left), or long double-stranded RNA (bottom left) which is often synthesized by an RNA-dependent RNA polymerase (RDR). Substrates are processed by Dicer-Like (DCL) proteins to yield initial duplexes. One of the two strands is bound to Argonaute (AGO) protein, and then guides the AGO protein to RNA targets based on complementarity.
B. Schematic of microRNA biogenesis from a hairpin precursor. MIRNA loci are defined as hairpins that produce a discrete initial duplex (the miRNA/miRNA* duplex). Red: miRNA, blue: miRNA*.
C. Schematic of small RNA biogenesis from a non-microRNA hairpin precursor, which is defined as small RNA-producing hairpin that does not meet the definition of a MIRNA locus.
D. Schematic of the biogenesis of secondary, phased siRNAs, which are defined by small RNA-directed cleavage of a long precursor, followed by dsRNA synthesis and dicing.
E. Schematic of heterochromatic siRNAs, which are defined by accumulation from intergenic regions and association with repressive DNA and histone modifications.
B-E: Modified from [60].