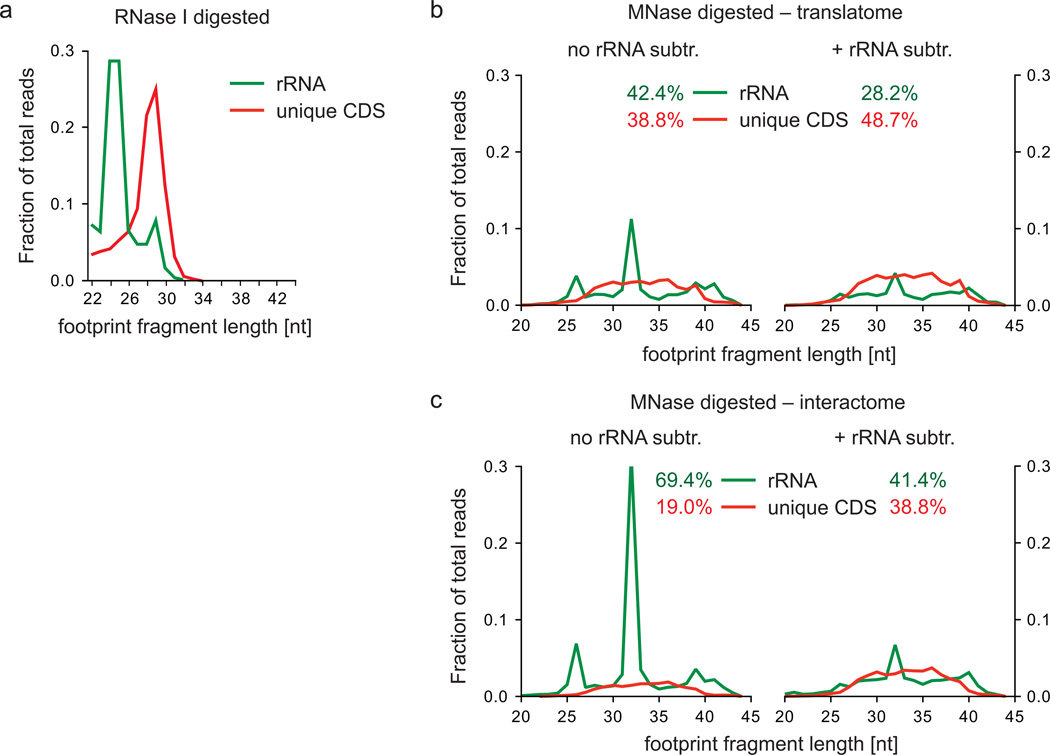
Figure 7.
Footprint fragment lengths and rRNA contamination variation according to digestion conditions. (a) Footprint fragments and rRNA contamination of RNase I digested yeast lysate (data from Ingolia et al.12) were plotted according to their lengths as fractions of total reads. (b,c) MNase-digested bacterial footprint fragments derived from translatome (b) or interactome (c) samples were prepared as described in the legend of Fig. 2 without (left panel) and after (right panel) rRNA depletion during the preparation of the deep sequencing library (steps 76-83 in the Supplementary Methods). Read lengths were plotted as fractions of total reads calculated according to step 41.