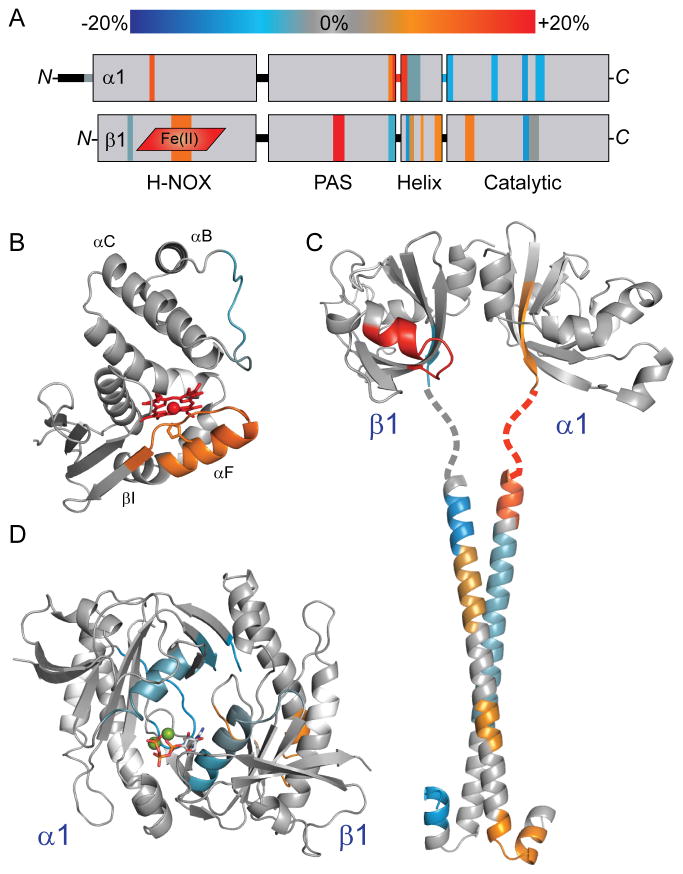
Figure 4. NO-induced conformational changes of full-length sGC mapped to domain structural models.
(A) Results from HDX-MS analysis of the NO-induced changes in α1β1 sGC (Figure 3) were condensed, color-coded according to the scale bar, and mapped to the representation of sGC domain organization.
(B) HD exchange rate perturbations induced by NO-binding were color-coded (as in Figure 3) and mapped to a structural model of the β1 H-NOX, as in Figure 2.
(C) NO-induced changes were mapped to a model of the α1β1 PAS-linker-helical domains. The α1 and β1 PAS domains were modeled independently using the Robetta server, based on homology with the M. sexta α1 PAS domain structure (PDB: 4GJ4) (Kim et al., 2004; Purohit et al., 2013). PAS domain monomers were dimerized by alignment with the structure of the bacterial PAS dimer (PDB: 2P04) (Ma et al., 2008). The structure of the rat sGC β1 helix (PDB: 3HLS) was used to reconstruct the coiled-coil of the helical domain (Ma et al., 2010). The α1 helical subunit was modeled using the Robetta server. The α1β1 coiled-coil was docked using the ClusPro 2.0 server, selecting the highest populated class exhibiting parallel orientation (Comeau et al., 2004). The intervening linker sequence is represented as a dotted line.
(D) NO-induced changes were mapped to the structure of the sGC α1β1 catalytic domain heterodimer (PDB: 3UVJ) (Allerston et al., 2013). For reference, GTP:Mg2+ substrate (sticks and balls) was docked into the active site.