Abstract
The scaffolding protein tetraspanin18 (Tspan18) maintains epithelial cadherin-6B (Cad6B) to antagonize chick cranial neural crest epithelial-to-mesenchymal transition (EMT). For migration to take place, Tspan18 must be downregulated. Here, we characterize the role of the winged-helix transcription factor FoxD3 in the control of Tspan18 expression. Although we previously found that Tspan18 mRNA persists several hours past the stage it would normally be downregulated in FoxD3-deficient neural folds, we now show that Tspan18 expression eventually declines. This indicates that while FoxD3 is crucial for initial downregulation of Tspan18, other factors subsequently impact Tspan18 expression. Remarkably, the classical EMT transcription factor Snail2 is not one of these factors. As in other vertebrates, FoxD3 is required for chick cranial neural crest specification and migration, however, FoxD3 has surprisingly little impact on chick cranial neural crest cell survival. Strikingly, Tspan18 knockdown rescues FoxD3-dependent neural crest migration defects, although neural crest specification is still deficient. This indicates that FoxD3 promotes cranial neural crest EMT by eliciting Tspan18 downregulation separable from its Tspan18-independent activity during neural crest specification and survival.
Keywords: neural crest, FoxD3, tetraspanin, migration, EMT
1. Introduction
The neural crest is a transient population of multipotent cells that arises from the dorsal neural tube of vertebrate embryos, migrates, and eventually contributes to a wide range of adult structures, including the peripheral nervous system and craniofacial skeleton (LeDouarin and Kalcheim, 1999). Neural crest development begins during gastrulation, when secreted factors induce the expression of a cohort of transcription factors, known generally as neural crest specifiers (Betancur et al., 2010), in the neural folds (Stuhlmiller and Garcia-Castro, 2012). Neural crest specifiers converge to regulate the activity of neural crest effector genes that control cell adhesion, motility, and fate (Betancur et al., 2010). By doing so, neural crest specifiers are crucial for neural crest cells to progress through epithelial-to-mesenchymal transition (EMT), migration, and differentiation into diverse adult neural crest derivatives. Unfortunately, since few targets of neural crest specifiers have been identified, we know little about the exact mechanism behind their cellular impacts.
The winged-helix transcription factor FoxD3 is a key neural crest specifier that has been implicated in multiple steps of neural crest development. FoxD3 is initially required for neural crest formation during early stages of neural crest development, including specification, multipotency, cell fate and survival (Kos et al., 2001; Sasai et al., 2001; Lister et al., 2006; Montero-Balaguer et al., 2006; Stewart et al., 2006; Thomas and Erickson, 2009; Mundell and Labosky, 2011; Hochgreb-Hagele and Bronner, 2013; Nitzan et al., 2013). Subsequently, neural crest cells fail to migrate in FoxD3 mutant zebrafish (Stewart et al., 2006). Furthermore, FoxD3 overexpression in the chick trunk neural tube alters cell-cell adhesion (Cheung et al, 2005) and increases the number of emigrating neural crest cells (Dottori et al., 2001; Kos et al., 2001). While these results suggest that FoxD3 also regulates neural crest migration, it is not possible to distinguish whether FoxD3 is required for neural crest migration independent of its functions during neural crest formation, or whether migration is blocked as a secondary consequence of these earlier roles. Moreover, FoxD3 represses neural crest and cancer cell migration in other circumstances, leaving the role of FoxD3 in migration unclear (Drerup et al., 2009; Katiyar and Aplin, 2011). Altogether, these results emphasize the need for further investigation to reconcile conflicting observations and define FoxD3 function during migration.
At the start of neural crest EMT, altered expression of Ca2+-dependent cell adhesion molecules called cadherins disrupts cell-cell adhesions (Meng and Takeichi, 2009; Nieto, 2011; Oda and Takeichi, 2011). In cranial neural crest cells, cadherin-6B (Cad6B) is directly repressed by the neural crest specifier and well-established EMT transcription factor, Snail2 (Coles et al., 2007; Taneyhill et al., 2007). However, Cad6B protein levels in cranial neural crest cells are not only subject to transcriptional control. ADAM metalloproteases cleave Cad6B, leading to its loss from the neural folds (Schiffmacher et al., 2014). Moreover, the transmembrane scaffolding protein Tetraspanin18 (Tspan18) post-translationally maintains Cad6B protein levels and must be downregulated in order for cranial neural crest cells to migrate (Fairchild and Gammill, 2013). Intriguingly, premigratory cranial neural crest cells lacking FoxD3 retain Tspan18 mRNA expression (Fairchild and Gammill, 2013). This finding, along with the observation that ectopic FoxD3 expression in chick trunk neural tube alters cell adhesion molecule expression (Cheung et al., 2005), suggests that FoxD3 may regulate migration by modulating cadherin levels during cranial neural crest EMT through its effects on Tspan18.
Although our previous study showed that FoxD3 knockdown sustained Tspan18 expression (Fairchild and Gammill, 2013), it did not address long-term outcomes. Moreover, when FoxD3 was knocked down, it was unclear whether Tspan18 expression persisted as an indirect consequence of altered neural crest specification, or whether Tspan18 was downstream of FoxD3 during EMT. Thus, the aim of this study was to distinguish between these two scenarios. We report that FoxD3 is required for initial downregulation of Tspan18, as well as for chick cranial neural crest specification and migration. Importantly, we show that Tspan18 knockdown rescues FoxD3-dependent migration but not specification defects. Altogether, these results indicate that FoxD3 promotes cranial neural crest migration at least in part through downregulation of Tspan18 independent of its role in other aspects of neural crest development.
2. Results
2.1. FoxD3 and Tspan18 expression overlap in premigratory cranial neural crest cells
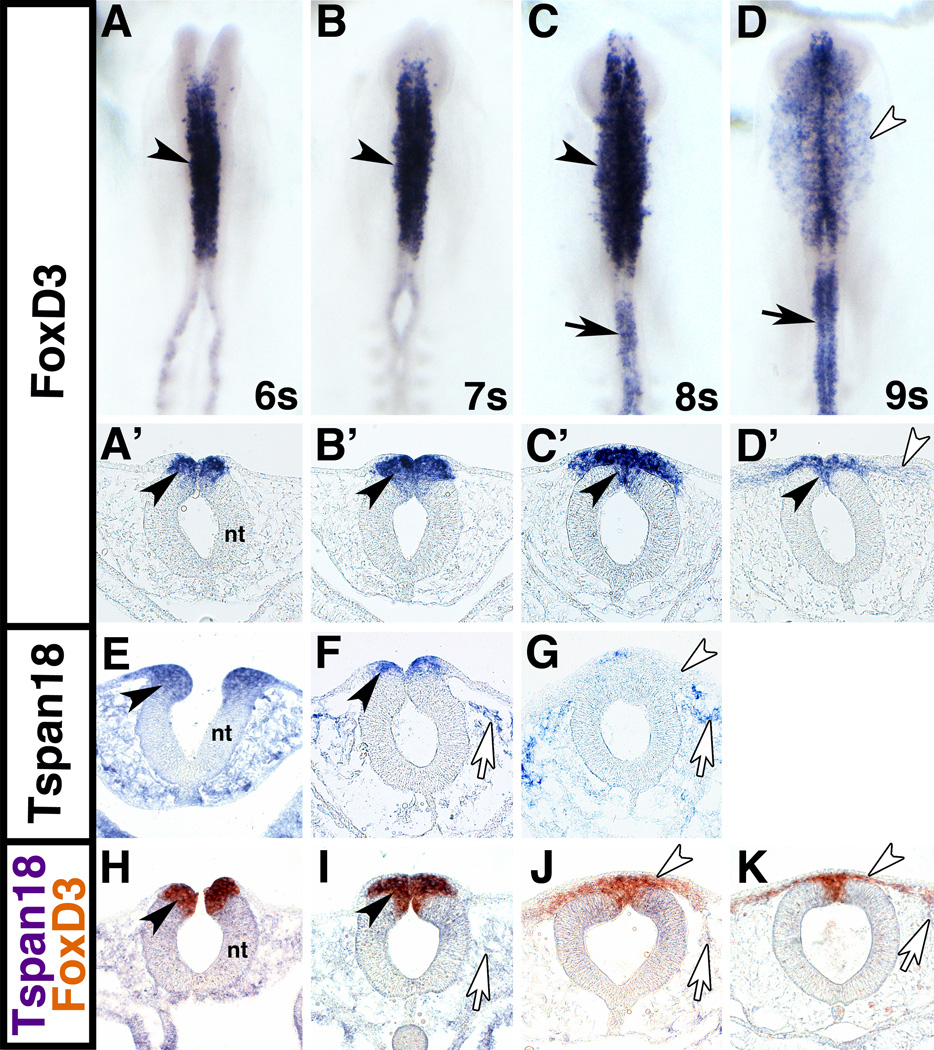
The general expression pattern of FoxD3 during chick cranial neural crest development has previously been described (Kos et al., 2001; Khudyakov and Bronner-Fraser, 2009; Simoes-Costa et al., 2012); however, for FoxD3 to regulate Tspan18, they must be co-expressed in neural crest cells as they are undergoing EMT, specifically before 8 somites when Tspan18 downregulation occurs (Fairchild and Gammill, 2013). To assess expression overlap, we visualized and compared FoxD3 and Tspan18 mRNA levels by in situ hybridization in whole mount and transverse sections. At 6 and 7 somites, FoxD3 transcripts were detected exclusively in the cranial neural tube (Fig. 1A,B, arrowheads). Transverse sections confirmed that FoxD3 was abundantly expressed in the dorsal neural tube at these stages (Fig. 1A’,B’). At 8s, emigrating cranial neural crest cells expressed FoxD3 mRNA, which persisted in the dorsal cranial neural tube (Fig. 1C,C’, arrowhead), and additionally extended into the trunk (Fig. 1C, black arrow). At 9s, FoxD3 expression was still apparent in the cranial dorsal neural tube (Fig. 1D’, black arrowhead) and in the trunk (Fig. 1D, black arrow); however, its expression was reduced in actively migrating neural crest cells (Fig. 1D,D’, white arrowheads). Likewise, Tspan18 mRNA expression in the cranial dorsal neural tube was apparent at 6 and 7 somites (Fig. 1E,F, black arrowheads), entirely overlapping with the FoxD3 expression domain (Fig. 1H,I, black arrowheads); however, Tspan18 expression was downregulated in the dorsal neural tube and migratory neural crest cells by 8s (Fig. 1G, white arrowhead). Neural crest cells migrating away from the neural tube expressed only FoxD3 (Fig. 1J, K, white arrowheads) and were surrounded by Tspan18 expression in the head mesenchyme (Fig. 1F,G,I-K white arrows; (Fairchild and Gammill, 2013)). Thus, FoxD3 is expressed at the right time and in the correct location to regulate Tspan18 expression.
Fig. 1. FoxD3 and Tspan18 are co-expressed in premigratory cranial neural crest cells.
Whole mount and transverse sections of chick embryos processed by in situ hybridization for FoxD3 (A-D), Tspan18 (E-G), or FoxD3 and Tspan18 (H-K) at 6 somites (s; A,A’,E, H), 7s (B,B’,F, I), 8s (C,C’,G, J) and 9s (D,D’, K). (A,B) FoxD3 (purple) is expressed in premigratory cranial neural crest cells at 6 and 7s (black arrowheads). (C) At 8s, FoxD3 expression persists in early emigrating neural crest cells (black arrowheads) and extends into the trunk (black arrow). (D) At 9s, FoxD3 expression is reduced in migrating cranial neural crest cells (white arrowhead), but retained in the dorsal neural tube (black arrowhead in D’) and in the trunk (arrow). (E-G) Tspan18 (purple) is expressed in premigratory cranial neural crest cells at 6 and 7s (black arrowheads), when its expression begins to downregulate. At 8s, Tspan18 is absent from migratory neural crest cells (white arrowhead). Tspan18 is expressed in scattered cells in the head mesenchyme (white arrows). (H-K) FoxD3 (orange) and Tspan18 (purple) expression domains overlap in the neural folds at 6s and 7s (black arrowheads). At 8s and 9s, migratory neural crest cells express FoxD3 but not Tspan18 (white arrowheads); Tspan18 expression is limited to head mesenchyme (white arrows). A-D, dorsal view; A’-D’, E-K, transverse sections. nt, neural tube.
2.2. In the absence of FoxD3, Tspan18 mRNA downregulation is delayed
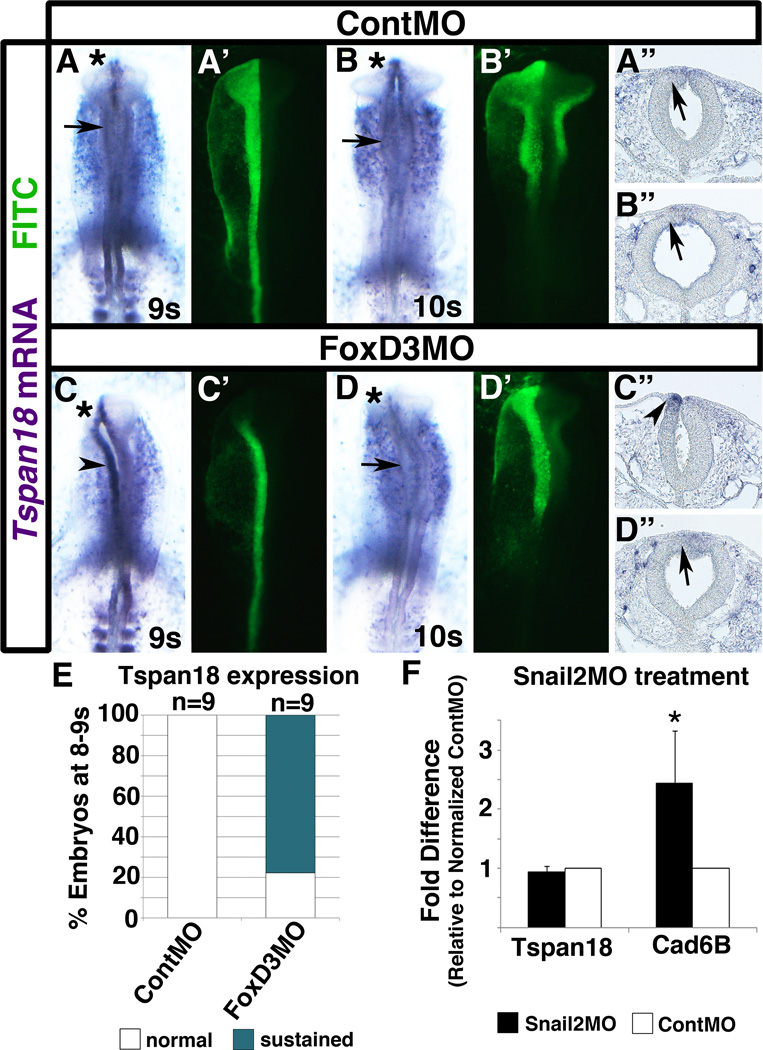
We previously reported that Tspan18 mRNA fails to downregulate when FoxD3 is knocked down (Fairchild and Gammill, 2013). To determine the persistence and dynamics of this effect, we evaluated Tspan18 expression over time in embryos electroporated with a FITC-tagged FoxD3 translation-blocking antisense morpholino oligonucleotide (FoxD3MO; (Kos et al., 2001)). FITC-tagged standard control MO (ContMO) or FoxD3MO was electroporated unilaterally into presumptive chick neural crest cells at stage HH4+, and resulting embryos at 8–9 or 10+ somites were processed by in situ hybridization to visualize Tspan18 mRNA expression in whole mount or transverse sections. As usual (Fig. 1; (Fairchild and Gammill, 2013)), Tspan18 mRNA was absent in the dorsal neural tube of embryos with 8 or more somites that had been electroporated with ContMO (Fig. 2A,A”,B,B”; arrows). In contrast, at 8–9 somites, Tspan18 mRNA persisted on the targeted side of the dorsal neural tube in embryos electroporated with FoxD3MO (Fig. 2C,C”,E; arrowheads). However, by 10 somites, Tspan18 transcripts were no longer visible in the dorsal neural tube of FoxD3MO-electroporated embryos (Fig. 2D,D”). These results suggest that FoxD3 is required for prompt, initial downregulation of Tspan18 mRNA, but it is not the only factor regulating Tspan18 mRNA expression.
Fig. 2. Tspan18 downregulation is initially delayed in the absence of FoxD3.
Whole mount images (A-D, A’-D’) and transverse sections (A”-D”) of embryos unilaterally electroporated with ContMO (A,B) or FoxD3MO (C,D) at late gastrula and processed by in situ hybridization for Tspan18 after harvest. At 9 somites (s; A) and 10s (B), Tpsan18 mRNA is absent (arrows) on the targeted (asterisks; green in A’,B’) and untargeted sides of the dorsal neural tube in representative ContMO-electroporated embryos (A,B) and transverse sections (A”,B”). At 9s, Tspan18 expression is retained (arrowhead) on the FoxD3MO-targeted side of the neural tube (asterisk; C’, green) in embryos (C) and transverse sections (C”). At 10s, Tspan18 mRNA expression is downregulated (arrow), even on the FoxD3MO-targeted side (asterisk; F’, green) of the neural tube in embryos (D) and transverse sections (D”). (E) Bar graph representing the frequency of 8–9s embryos exhibiting sustained Tspan18 expression. (F) Quantification of Tspan18 mRNA levels following MO-mediated depletion of Snail2. Cad6B levels were analyzed as a positive control (Taneyhill et al., 2007). Results are reported as fold difference relative to that obtained with ContMO normalized to 1. Means and standard errors of fold differences were generated from 3 independent experimental cDNA replicates. The asterisk (*) located above the error bar denotes a significant difference in the fold change between Snail2MO and ContMO levels (p< 0.05). Black bars, Snail2 MO; white bars, ContMO.
One obvious candidate to contribute to Tspan18 downregulation is the neural crest transcriptional repressor, Snail2. Tspan18 antagonizes EMT by maintaining Cad6B protein (Fairchild and Gammill, 2013), and Cad6B is directly repressed by Snail2 (Taneyhill et al., 2007). Thus, we reasoned that Tspan18 might also be regulated by Snail2. To determine if Snail2 directly regulates Tspan18 in cranial neural crest cells, we electroporated 5 to 6 somite embryos with either ContMO or Snail2MO, excised MO-targeted neural folds after 30 min, and quantified Cad6B (as a positive control) and Tspan18 mRNA levels by QPCR. As expected, Cad6B mRNA levels were significantly increased in Snail2MO-electroporated embryos as compared to ContMO-electroporated embryos (Fig. 2F; approximately 2.5-fold increase, p<0.05, as previously described; (Taneyhill et al., 2007)), suggesting that Snail2MO was effective and efficiently reduced Snail2 protein levels. In contrast, Tspan18 mRNA levels were similar in both Snail2MO- and ContMO-electroporated embryos (Fig. 2F). Thus, Snail2 is not one of the additional transcription factors that regulate Tspan18 expression. The identity of these additional factors awaits further analysis.
2.3. FoxD3 minimally impacts cranial neural crest cell survival
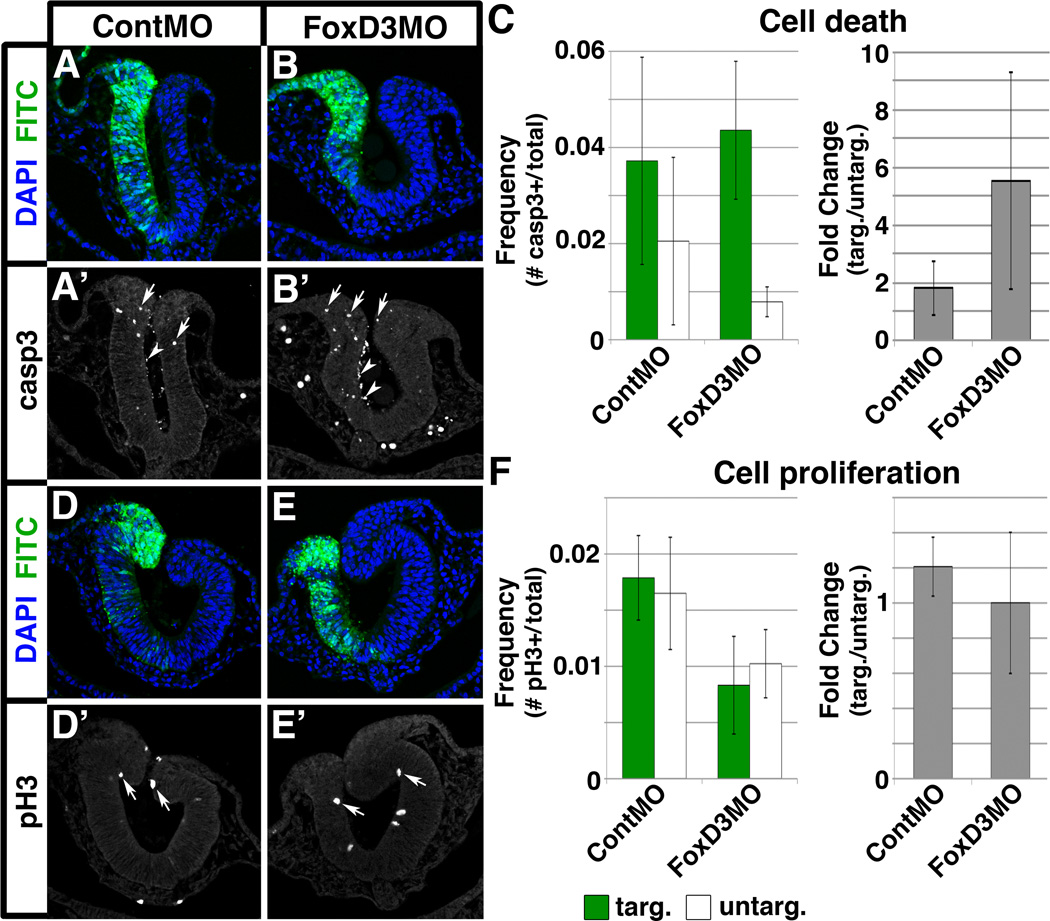
Because FoxD3 is required for neural crest cell survival in other systems (Stewart et al., 2006; Wang et al., 2011), we next determined whether FoxD3 knockdown altered cell death in chick cranial neural crest cells. To visualize dying premigratory neural crest cells, we immunostained FoxD3MO-electroporated embryos for the activated, cleaved form of the apoptotic enzyme caspase-3 (casp3) and determined the frequency of casp3-positive cells out of the total number of cells in the dorsal neural tube. While we electroporated FoxD3MO at 1.0 mM previously (Fairchild and Gammill, 2013), cells electroporated with FoxD3MO at 1.0 mM were strongly and non-specifically apoptotic (C. Fairchild, unpublished). Electroporation of 0.5 mM FoxD3MO largely prevented this response, so that very few dying cells were apparent in the neural tube ventral to the FoxD3 expression domain (Fig. 3A’,B’; both control- and FoxD3MO-electroporated embryos exhibited casp3-positive cells in the lumen of the neural tube (arrowheads) that were not counted during quantification). In 0.5 mM FoxD3MO-electroporated embryos, the frequency of casp3-positive cells in the dorsal neural tube was significantly higher on the targeted side compared to the untargeted side (Fig. 3B,B’,C; p=0.003). However, the absolute number of dying cells was small (averaging 2.7 ContMO-electroporated cells and 2.5 FoxD3MO-electroporated cells per dorsal neural tube), with ContMO- and FoxD3MO-targeted sides exhibiting a similar, low frequency of casp3-positive cells (roughly 1–6% of cells counted; Fig. 3C, compare green bars). As a result, the fold change of the targeted compared to untargeted side of the dorsal neural tube in ContMO- and FoxD3MO-electroporated embryos was not significantly different (Fig. 3C), suggesting that reduced cell survival is a minor factor in the FoxD3 knockdown phenotype. To ensure there was no reciprocal effect on proliferation, we immunostained for phospho-histone H3 (pH3) and found that loss of FoxD3 did not alter the frequency of proliferating cells (Fig. 3D–F). In summary, while FoxD3 knockdown does slightly increase the frequency of dying cells, consistent with results in zebrafish (Stewart et al., 2006; Wang et al., 2011), it does not affect the overall fold-change in cell death. Moreover, loss of a small number of dying cells in the dorsal neural tube cannot explain the maintenance of Tspan18 mRNA expression throughout the neural fold (Fig. 2). Thus, we conclude that FoxD3 has a nominal role in chick cranial neural crest cell survival that is independent of its role in regulating Tspan18 mRNA expression.
Fig. 3. FoxD3 knockdown has minimal effects on cell survival and does not alter cell proliferation.
Quantification of cell death (A-C) and proliferation (D-F) in embryos unilaterally electroporated with ContMO (A,D) or FoxD3MO (B,E). (A,B) Immunostaining for cleaved caspase-3 (casp3) reveals more dying cells (arrows) on the targeted side of the dorsal neural tube (green) in FoxD3MO-electroporated embryos (B) as compared to the targeted side of control embryos (A). Casp3-positive cells were present in the lumen of the neural tube (arrowheads) under both conditions and were thus not counted during quantification. (C) Bar graphs representing the frequency and fold change of dying cells in electroporated embryos. (D,E) Immunostaining for phospho-histone H3 (pH3) reveals that proliferating cells are equally apparent (arrows) on the targeted (green) and untargeted sides of the neural tube in embryos electroporated with either ContMO (D) or FoxD3MO (E). (F) Bar graphs representing the frequency and fold change of proliferating cells in electroporated embryos.
2.4. FoxD3 is required for premigratory Sox10 expression and neural crest migration
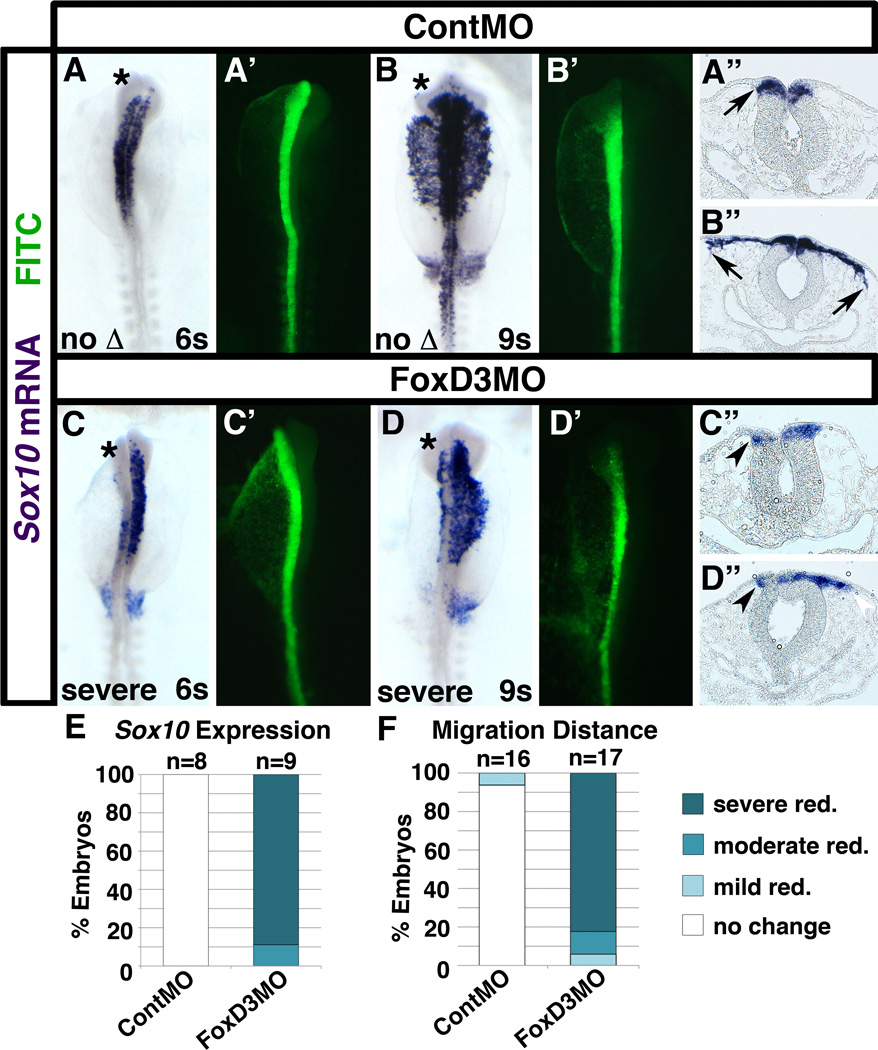
FoxD3 knockdown leads to transiently sustained Tspan18 mRNA expression (Fig. 2). Our previous report concluded that downregulation of Tspan18 is a requirement for neural crest cells to migrate (Fairchild and Gammill, 2013). To interpret the effects of FoxD3 knockdown in the context of early neural crest development, we electroporated stage HH4+ embryos with ContMO or FoxD3MO and visualized neural crest specification and migration by in situ hybridization for the key neural crest transcription factor and FoxD3-responsive gene, Sox10 (Prasad et al., 2012). Sox10 mRNA expression levels were unaffected at 6–7 somites in ContMO electroporated embryos (Fig. 4A,A”; arrow). However, Sox10 mRNA expression was severely reduced on the targeted side of the neural tube in embryos electroporated with FoxD3MO (Fig. 4C,C”,E; arrowhead), suggesting that cranial neural crest specification is inhibited by FoxD3 knockdown in chick embryos. This is consistent with analyses of mutant mouse and zebrafish embryos, which show that FoxD3 is required for early neural crest specification and precursor maintenance (Montero-Balaguer et al., 2006; Stewart et al., 2006; Teng et al., 2008; Wang et al., 2011; Hochgreb-Hagele and Bronner, 2013). Furthermore, the FoxD3-deficient Sox10-positive neural crest cells that did form failed to migrate. While ContMO-targeted and untargeted Sox10-positive neural crest cells migrated an equivalent distance away from the neural tube at 9–10 somites (Fig. 4B,B”; arrows), FoxD3MO-electroporated neural crest cell migration distance was severely reduced relative to the untargeted side in the majority of embryos (Fig. 4D,D”,F; arrowhead). These results indicate that FoxD3 is required for chick cranial neural crest formation (specification and, to a minor extent, survival) as well as subsequent migration. However, as in previous studies of FoxD3, these results do not distinguish whether FoxD3 is required for neural crest migration, or whether migration defects are an indirect consequence of impaired neural crest specification.
Fig. 4. Chick FoxD3 is required for premigratory Sox10 expression and neural crest migration.
Whole mount images (A-D, A’-D’) and transverse sections (A”-D”) of embryos unilaterally electroporated with ContMO (A,B) or FoxD3MO (C,D) at late gastrula, incubated to 6 somites (s; A,C) or 9s (B,D), and processed by in situ hybridization for Sox10. In ContMO-electroporated embryos, Sox10 expression (A,A”) and the distance that Sox10-positive neural crest cells migrated (B,B”) was even on targeted (asterisks; A’,B’, green) and untargeted sides of the neural tube. In contrast, Sox10 expression was impaired (C,C”) and migration distance was reduced (D,D”) on the targeted side (asterisks; C’,D’, green) compared to the untargeted side of the neural tube of FoxD3MO-electroporated embryos. (E,F) Bar graphs representing the frequency of electroporated embryos exhibiting reduced premigratory Sox10 expression (E) or reduced migration distance (F).
2.5. FoxD3 regulates cranial neural crest migration through Tspan18
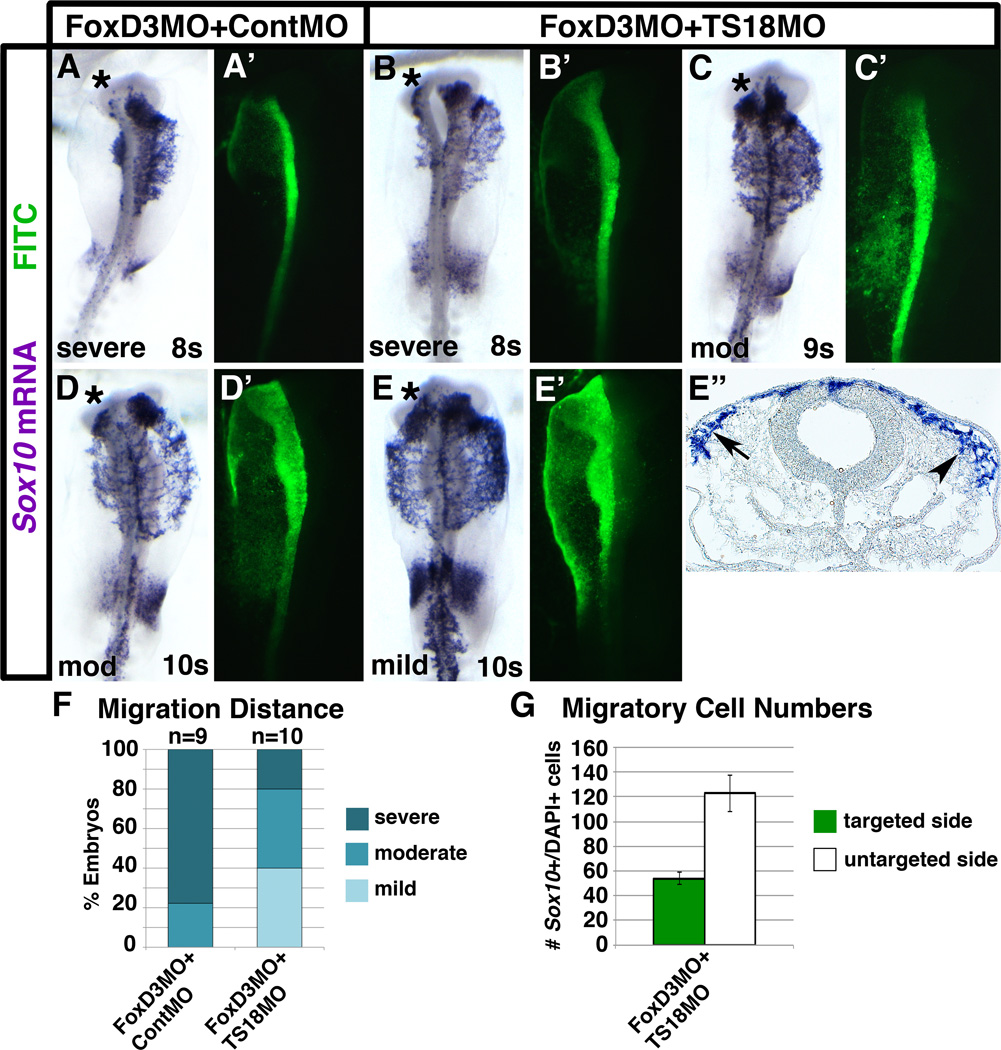
Given that expression of Tspan18 is incompatible with migration (Fairchild and Gammill, 2013), it is possible that cranial neural crest cells fail to migrate in FoxD3 knockdown embryos (Fig. 4) because they retain Tspan18 expression (Fig. 2; (Fairchild and Gammill, 2013)). If this were true, we reasoned that knockdown of Tspan18 should rescue the FoxD3 loss-of-function migration phenotype. To investigate this possibility, we co-electroporated embryos with FoxD3MO plus either ContMO or TS18MO, processed the resulting embryos by in situ hybridization for Sox10 and scored the distance Sox10-positive neural crest cells had migrated away from the neural tube. Similar to embryos electroporated with FoxD3MO alone (Fig. 4D), in embryos co-electroporated with FoxD3MO + ContMO, neural crest migration distance was severely reduced on the targeted side of the neural tube in 7/9 (Fig. 5A,F) and moderately reduced in 2/9 (Fig.5D,F). Strikingly, in embryos co-electroporated with FoxD3MO + TS18MO, only 2/10 exhibited severe migration defects (Fig.5B,F), 3/10 were moderately reduced (Fig. 5C,F), and neural crest migration in 4/10 embryos was only mildly affected (Fig. 5E,F). Fluorescence imaging verified that rescued migratory neural crest cells were FITC (MO)-positive (Fig. 5B’,C’,E’ and data not shown). Thus, knockdown of Tspan18 partially rescues the FoxD3 loss-of-function migration phenotype. Given that Tspan18 antagonizes EMT by maintaining Cad6B protein (Fairchild and Gammill, 2013), this suggests that FoxD3 promotes cranial neural crest EMT and thus migration by eliciting Tspan18 downregulation.
Fig. 5. Tspan18 knock down rescues FoxD3-dependent migration defects.
Whole mount images and transverse sections of embryos unilaterally co-electroporated with FoxD3MO and either ContMO (A,D) or TS18MO (B,C,E) at late gastrula, incubated to 8–10 somites (s), and processed by in situ hybridization for Sox10. (A-E) Following co-electroporation with FoxD3MO and ContMO, the distance of Sox10-positive cells migrated on the targeted side of the neural tube (asterisks; green in A’-E’) is severely inhibited (A) in nearly all embryos with only a few moderately affected (D). Following co-electroporation with FoxD3 and TS18MO, the incidence of severely affected (B) embryos is greatly reduced and most embryos are moderately (mod; C) or only mildly affected (E). (E”) A transverse section of the mildly affected embryo in (E) reveals that while co-electroporation with FoxD3MO and TS18MO rescues the emigration of migratory neural crest cells, the size of the Sox10-positive migratory neural crest population is reduced (arrow) on the targeted side of the embryo, as compared to the untargeted side of the embryo (arrowhead). (F) Bar graph representing the frequency of electroporated embryos that exhibit a severe, moderate, or mild reduction in migration distance. (G) Bar graph showing the reduced number of Sox10-positive, DAPI-positive cells adjacent to the neural tube on the targeted (green) versus untargeted (white) side of embryos electroporated with FoxD3MO and TS18MO.
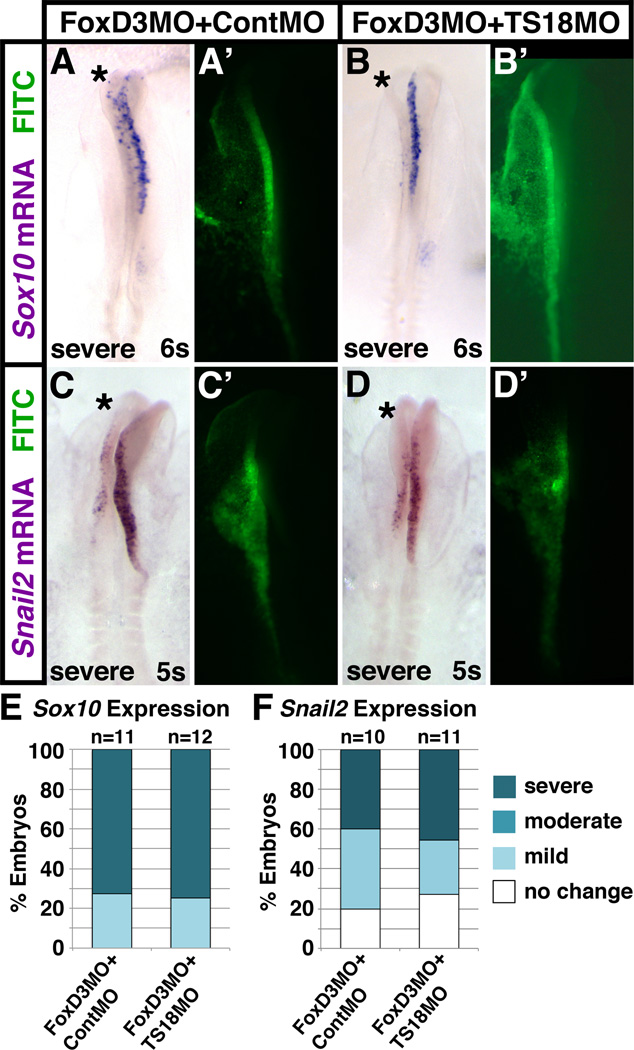
While scoring the rescued embryos, we noted that, although co-electroporation of TS18MO with FoxD3MO rescued migration, the migratory population was smaller (see for example Fig. 5E”, arrow). Indeed, counting Sox10-positive cells adjacent to the FoxD3MO + TS18MO targeted and untargeted sides of the neural tube showed that there were 2.3-fold fewer migratory neural crest cells on the targeted side (Fig. 5G). One possible explanation for this could be that Tspan18 knockdown does not rescue FoxD3-dependent specification defects. To test this hypothesis, we once again co-electroporated embryos with FoxD3MO and either ContMO or TS18MO, but instead harvested embryos at 4–7 somites and assayed Sox10 and Snail2 expression by in situ hybridization. Compared to the untargeted side, Sox10 (Fig. 6A,E) and Snail2 (Fig. 6C, F) expression were severely reduced on the FoxD3MO + ContMO targeted side of the neural tube. Importantly, Sox10 (Fig. 6B,E) and Snail2 (Fig. 6D,F) expression were similarly affected by co-electroporation of FoxD3MO + TS18MO. Therefore, Tspan18 knockdown does not rescue FoxD3-dependent neural crest specification. Thus, when incubated to migration stages, neural crest cells deficient for FoxD3 and Tspan18 are able to undergo EMT and leave the neural tube, but fewer are present to do so. In sum, these results reveal that FoxD3 regulates cranial neural crest migration through its effects on Tspan18 expression. Moreover, the failure of Tspan18 knockdown to rescue FoxD3-dependent cranial neural crest gene expression indicates that FoxD3 separably regulates cranial neural crest specification and migration through Tspan18-independent and -dependent pathways (respectively).
Fig. 6. FoxD3 is required for neural crest specification independent of Tspan18.
Whole mount images of embryos unilaterally co-electroporated with FoxD3MO and either ContMO (A,C) or TS18MO (B,D) at late gastrula, incubated to 5–6 somites (s), and processed by in situ hybridization for Sox10 (A,B) or Snail2 (C,D). (A-D) Co-electroporation of FoxD3MO with either ContMO or TS18MO disrupts Sox10 (A,B) and Snail2 (C,D) expression on the targeted side of the neural tube (asterisks; green in A’-D’). (E,F) Bar graphs representing the frequency of electroporated embryos that exhibit a severe, moderate, mild, or no reduction in Sox10 (E) or Snail2 (F) expression, indicating that Tspan18 knockdown has no impact on the severity of FoxD3-dependent specification defects.
3. Discussion
In this study, we have investigated the regulatory relationship between FoxD3 and Tspan18 and, in so doing, gained insight into the poorly understood role of FoxD3 in neural crest migration. From our FoxD3 knockdown experiments, we conclude that FoxD3 supports chick cranial neural crest specification and migration, minimally affects neural crest cell survival, and specifically report that FoxD3 is required for initial downregulation of Tspan18 mRNA expression. In fact, FoxD3 regulates neural crest migration through its effects on Tspan18, as FoxD3-dependent migration defects can be rescued by simultaneous knockdown of Tspan18, although neural crest specification is still aberrant. Together these data reveal separable requirements for FoxD3 during cranial neural crest specification and migration, and show that FoxD3 promotes EMT and migration through negative regulation of Tspan18 expression.
3.1. FoxD3 is required for initial Tspan18 downregulation, though other factors contribute at later stages
Tspan18 expression (overexpression or persistent endogenous transcripts) is incompatible with neural crest migration (Fairchild and Gammill, 2013), and as such, Tspan18 mRNA is absent from cranial neural crest cells later than 7 somites (Fig. 1). In embryos electroporated with FoxD3MO, Tspan18 transcripts are still visible at 9 somites (Fig. 2C), indicating that FoxD3 is required for the initial downregulation of Tspan18. Although the Tspan18 promoter contains FoxD3 consensus binding sites, we have, to date, been unable to determine whether FoxD3 directly binds and represses Tspan18 transcription. Experiments to address this crucial question will be the subject of future investigation.
Our results highlight the complexity of Tspan18 transcriptional regulation. Notably, even in embryos that have been electroporated with FoxD3MO, Tspan18 mRNA is eventually downregulated (Fig. 2D). This finding suggests that FoxD3 is not the only factor that negatively regulates Tspan18 mRNA expression. The identity of these additional factors is unclear, though Snail2 does not appear to contribute (Fig. 2F). Regulatory complexity is further indicated by prolonged co-expression of FoxD3 and Tspan18 in premigratory neural crest cells (Fig. 1). This delayed action of a neural crest transcription factor is not unprecedented: Snail2, which represses Cad6B transcription to promote EMT, is co-expressed with Cad6B throughout premigratory neural crest development and modulated through altered rates of degradation (Vernon and LaBonne, 2006; Taneyhill et al., 2007). FoxD3 activity could also undergo differential post-translational modifications, as does Sox10 (Taylor and Labonne, 2005). Alternatively, FoxD3 could require and/or affect the activity of a co-factor, for which there is also precedent. In the trunk neural crest, FoxD3 regulates the expression of the melanocyte marker MITF, but not through direct binding to the MITF promoter; instead FoxD3 binds to the transcriptional activator Pax3 and prevents Pax3 from binding to the MITF promoter (Thomas and Erickson, 2009). Identifying FoxD3-interactors would provide a better understanding of its regulation of Tspan18 mRNA expression.
3.2 FoxD3 promotes Tspan18 downregulation to drive cranial neural crest EMT independent of its role in specification
The ability of Tspan18 to rescue FoxD3-dependent migration (Fig. 5) allowed us to separate the requirement for FoxD3 during neural crest formation and migration. Consistent with previous studies in Xenopus, zebrafish and mouse embryos (Sasai et al., 2001; Lister et al., 2006; Montero-Balaguer et al., 2006; Stewart et al., 2006; Mundell and Labosky, 2011; Hochgreb-Hagele and Bronner, 2013), FoxD3 knockdown in chick embryos inhibited neural crest specification (Fig. 4C). While FoxD3 is also necessary for neural crest migration (Fig. 4D; (Stewart et al., 2006)), it had been unclear whether FoxD3 regulates neural crest migration directly, or affects migration as a secondary consequence of specification and survival defects. Crucially, co-electroporation of TS18MO with FoxD3MO rescues the ability of neural crest cells to migrate (Fig. 5B,C,E,F), but not the essential role of FoxD3 during neural crest specification (Fig. 6B,D-F). In other words, fewer neural crest cells form when FoxD3 is deficient (Fig. 5G), but those that arise will emigrate as long as Tspan18 is knocked down. This indicates that FoxD3 regulates neural crest EMT/migration through downregulation of Tspan18 and provides the first evidence that FoxD3 plays independent roles in neural crest specification and migration (Fig. 7). FoxD3 knockdown only nominally affected survival in chick embryos (Fig.3A–C), and because TS18MO does not affect cell death or proliferation (Fairchild and Gammill, 2013), FoxD3/Tspan18-dependent migration defects are likely to independent of cell death as well.
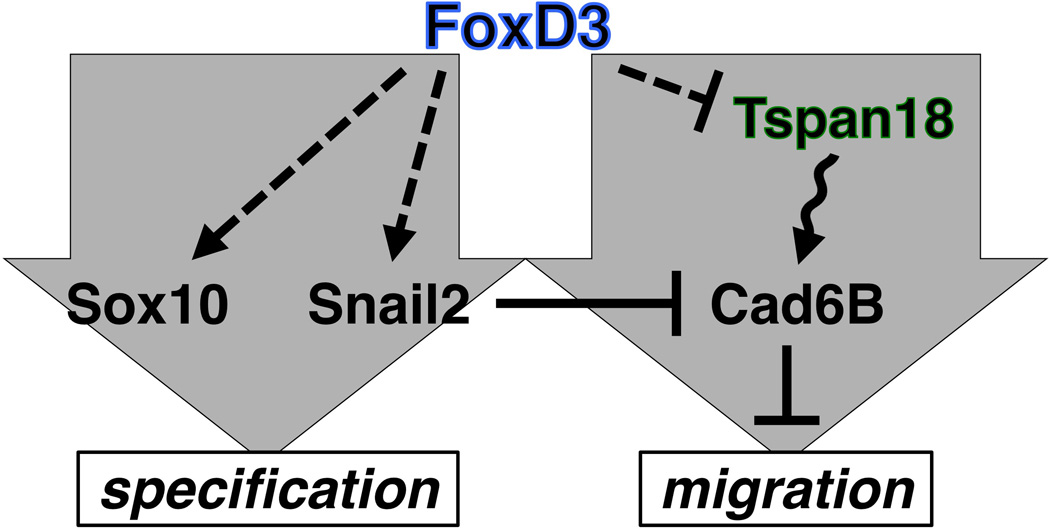
Fig. 7. FoxD3 independently regulates neural crest specification and migration.
FoxD3 is required for Sox10 and Snail2 expression (Figs. 4, 6) in the gene regulatory network that drives neural crest specification (Betancur et al., 2010; Prasad et al., 2012). In a separate pathway (grey arrow), FoxD3 promotes neural crest migration through its effects on Tspan18 (Fig. 5), repressing Tspan18 expression (Fig. 2) and relieving Tspan18-dependent maintenance of Cad6B (Fairchild and Gammill, 2013) that prevents EMT (Coles et al., 2007). Snail2 likewise promotes EMT by directly repressing Cad6B (Taneyhill et al., 2007) but does not affect Tspan18 expression (Fig. 2). Grey arrows, pathways; arrowhead, promotes expression; bar, represses expression; solid line, direct binding to target promoter demonstrated; dashed line, regulation not necessarily direct; wavy line, post-translational effect; box, developmental outcome.
As FoxD3 has been reported to both positively and negatively regulate neural crest migration, we propose that FoxD3 promotes EMT but is incompatible with motility. FoxD3 is required for neural crest migration (Fig. 4,6; (Stewart et al., 2006)). However, expanded FoxD3 expression in zebrafish cranial neural crest cells prevents migration (Drerup et al., 2009), and FoxD3 negatively regulates Rho-GTPase-induced cell motility in human metastatic melanoma cells (Katiyar and Aplin, 2011). These contrary observations likely reflect temporal differences. FoxD3 knockdown sustains Tspan18 expression (Fig. 2), which in turn maintains Cad6B protein levels that inhibit neural crest EMT (Coles et al., 2007; Fairchild and Gammill, 2013). Thus, at the start of neural crest migration, FoxD3 elicits Tspan18 downregulation, which destabilizes Cad6B-dependent cell adhesions and promotes neural crest EMT (Fig. 7). Later, in actively migrating neural crest cells, FoxD3 may disrupt Rho-based motility, as has been reported for melanoma cells (Katiyar and Aplin, 2011). This could explain why FoxD3 expression declines after neural crest cells leave the neural tube (Fig. 1; (Khudyakov and Bronner-Fraser, 2009)).
While FoxD3 regulates cranial neural crest EMT through Tspan18, it must act through a different pathway in the trunk. Although Tspan18 is not detectable by in situ hybridization in trunk neural tube (Fairchild and Gammill, 2013), FoxD3 overexpression leads to altered expression of cell adhesion molecules (Cheung et al., 2005). Thus, FoxD3 must regulate the expression of other gene(s) that impact EMT. To understand the role of FoxD3 in neural crest migration, and specifically in EMT, it will be crucial to identify FoxD3 targets in the neural crest.
In conclusion, this report shows that FoxD3 is required for chick cranial neural crest migration because it promotes the downregulation of Tspan18 and thus EMT. Moreover, this pathway is independent of the requirement for FoxD3 during cranial neural crest specification (Fig. 7). Overall this study provides new insight into the dual role of FoxD3 for neural crest formation and migration, and also gives us a better understanding for how neural crest transcription factors impact the cellular behaviors that occur as neural crest cells undergo EMT and migrate.
4. Experimental Procedures
4.1. Embryos
Fertile chicken embryos were incubated in a humidified incubator (G. Q. F. Manufacturing: Savannah, GA) at 37–38°C. Embryos were staged according to Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1951) or by counting somite pairs.
4.2. In situ hybridization
Whole mount in situ hybridization was performed as previously described (Wilkinson, 1992). Digoxigenin-labeled RNA probes were transcribed from the following templates: FoxD3 (Kos et al., 2001), Tspan18 (Gammill and Bronner-Fraser, 2002), and Sox10 (Cheng et al., 2000). For double whole mount in situ hybridization, RNA probes labeled with digoxygenin (Tspan18) and FITC (FoxD3) were mixed during hybridization. After developing the digoxygenin probe, alkaline phosphatase was inactivated by incubating 1 hour in 4% paraformaldehyde + 0.1% glutaraldehyde and 20 minutes at 63°C in maleic acid buffer + 0.1% Tween and 10mM EDTA. Orange precipitate was generated by incubating in 87.5 µg/ml iodonitrotetrazolium chloride (Sigma Aldrich; St Louis, MO) plus 87.5 µg/ml 5-bromo-4-chloro-3-indolyl-phosphate (Roche Applied Science; Indianapolis, IN). After processing, embryos were first imaged in whole mount using a Zeiss Discovery V8 stereoscope then infiltrated with 5% and 15% sucrose, embedded in gelatin, sectioned using a Leica CM1900 cryostat at 12–18 µm and imaged again using a Zeiss AxioImager A1. Images of whole mount embryos and transverse sections were taken with an AxioCam MRc5 digital camera and Axiovision software and assembled in Photoshop (Adobe).
4.3. Morpholino design and electroporation
The following FITC-tagged, antisense morpholinos (MO) were designed and synthesized by GeneTools, LLC (Philomath, OR): FoxD3 translation blocking MO (FoxD3MO: 5’-CGCTGCCGCCGCCCGATAGAGTCAT-3’; (Kos et al., 2001)), Tspan18 translation blocking MO (TS18MO: 5’-TGCAGCTCAGACAGTCTCCCTCCAT-3’; (Fairchild and Gammill, 2013)), and standard FITC control MO (ContMO: 5’-CCTCTTACCTCAGTTACAATTTATA-3’). For early embryo electroporations, morpholinos were unilaterally electroporated into the presumptive neural crest at HH stage 4+, as previously described (Gammill and Krull, 2011), at 500 µM (FoxD3 and ContMO) and 390 µM (TS18MO) with carrier DNA (0.2 µg/µL pCS2+MycTag DNA). After electroporation, embryos were re-incubated until the desired stages and fixed in 4% paraformaldehyde at room temperature for 1 hour then washed with PBS + 0.1% Tween. Targeting was verified by fluorescent microscopy for FITC before embryos were either immediately embedded and sectioned for immunofluorescence or dehydrated into methanol and stored at −20° for in situ hybridization.
4.4. Quantitative polymerase chain reaction (QPCR)
MO-mediated Snail2 depletion followed by QPCR was carried out as described (Taneyhill et al., 2007). Briefly, 6 somite embryos were electroporated in ovo with Snail2MO or ContMO and midbrain neural folds were excised after 30 minutes. Neural folds from six electroporated embryos were pooled and total RNA was isolated using the RNAqueous Total RNA Isolation Kit (Ambion-Life Technologies; Carlsbad, CA). cDNA was synthesized using random hexamers and the Superscript II RT-PCR system (Life technologies) according to the manufacturer’s instructions. QPCR was performed using the ABI 7000 in a TaqMan or SYBR Green (Life Technologies) assay as described (Taneyhill and Bronner-Fraser, 2005). Briefly, 25 µl Tspan18 QPCR reactions included 2X SYBR Green master mix, cDNA and 75 nM of each primer (Sense: 5´-GCTTGTTGCCAGCGAAAGCTCC-3´; Antisense: 5´-TAGCAGCCCTGCCGGTTCTGA-3´). 18S QPCR reactions included 2x SYBR Green mastermix, cDNA and 150 nM each primer as previously described (Taneyhill and Bronner-Fraser, 2005). Cad6B QPCR reactions included TaqMan mastermix, cDNA, 150 nM of each primer, and 150 nM each Cad6B TaqMan probe, as described (Taneyhill et al., 2007). After normalization to a standard (chick 18S rRNA), fold upregulation or downregulation for each of three replicates was determined by dividing the relative expression value for the gene of interest in the presence of the Snail2MO by that obtained for the gene of interest in the presence of ContMO.
4.5. Immunofluorescence and cell death/proliferation
Immunofluorescence was performed using either anti-cleaved caspase3 (casp3, Cell Signaling; Danvers, MD: 1:200) or anti-phosho-histone H3 (pH3, Millipore; Billerica, MA: 1:250) primary antibody diluted into PBS + 0.1% Triton-X100 supplemented with 5% normal donkey serum followed by incubation with a donkey anti-rabbit secondary antibody at 1:250 (RRX conjugated; Jackson Labs; West Grove, PA). Slides were mounted in PermaFluor (Thermo Fisher Scientific; Waltham, MA) containing 1 µg/mL DAPI and imaged on a Zeiss LSM 710 laser scanning confocal microscope. Images were assembled in Photoshop (Adobe). Cell death and proliferation were quantified as follows: casp3- or pH3-positive cells and DAPI-positive cells were counted in the dorsal quarter of the neural tube on the targeted and untargeted sides in at least 5 transverse sections per embryo (n=3 embryos). Frequency was determined by dividing the number of casp3- or pH3-positive cells by the total number of DAPI-positive cells, and fold change determined by dividing by the frequency on the targeted by the untargeted side. Statistics were performed using the paired Student’s t test in Excel (Microsoft).
4.6. Assaying neural crest migration
To consistently assay neural crest cell migration (Fig. 4) and thus clearly determine rescue (Fig. 5), the farthest distance Sox10-positive cells had migrated from the midline in whole mount was measured in Photoshop (Adobe) on the untargeted and targeted sides of the neural tube. The ratio of targeted distance / untargeted distance was then used to categorize phenotypic severity. In severely affected embryos, neural crest cells on the targeted side had migrated 0–50 percent the distance of those on the untargeted side. In moderately affected embryos, neural crest cells on the targeted side had migrated 51–75 percent of the distance of those on the untargeted side. In mildly affected embryos, neural crest cells on the targeted side had migrated 76–95 percent of the distance of those on the untargeted side. Unaffected embryos displayed neural crest migration distances within 5 percent of each other on the targeted and untargeted sides of the neural tube. To quantify effects on migratory cell number, Sox10 and DAPI images of 5 transverse sections from each of 4 embryos (20 sections total) were superimposed in Photoshop (Adobe). For each image, an outline was drawn around all Sox10 staining outside of the neural tube, and DAPI-positive nuclei within this outline were counted. Statistics were performed using the paired Student’s t test in Excel (Microsoft).
Highlights.
Factors in addition to FoxD3, but not Snail2, contribute to Tspan18 downregulation.
Chick cranial neural crest specification and migration require FoxD3.
FoxD3 promotes cranial neural crest migration by downregulating Tspan18.
During neural crest specification, the role of FoxD3 is Tspan18-independent.
FoxD3 separably regulates neural crest formation and migration.
Acknowledgements
We thank Yi-Chuan Cheng and Carol Erickson for the kind gift of plasmids. Many thanks to the members of the Gammill Lab for their input and support. We are grateful for the resources of the University of Minnesota Supercomputing Institute. This work was supported by the National Institutes of Health, award numbers F31 NRSA GM087951 (CLF), R00 HD055034 (LAT), and K22 DE015309 (LSG), as well as a University of Minnesota Grant-in-Aid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
L.S.G and C.L.F conceived and designed the experiments. C.L.F. and J.P.C. performed the electroporation and in situ hybridization experiments. A.T.S. and L.A.T. performed the QPCR experiments and analyzed those data. L.S.G. and C.L.F. analyzed the remaining data, and L.S.G. and C.L.F. wrote the paper with input from L.A.T.
References Cited
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res Dev Brain Res. 2000;121:233–241. doi: 10.1016/s0165-3806(00)00049-3. [DOI] [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–4138. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136:2623–2632. doi: 10.1242/dev.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CL, Gammill LS. Tetraspanin18 is a FoxD3-responsive antagonist of cranial neural crest epithelial to mesenchymal transition that maintains Cadherin6B protein. J Cell Sci. 2013;126:1464–1476. doi: 10.1242/jcs.120915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–5741. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Krull CE. Embryological and genetic manipulation of chick development. Methods Mol Biol. 2011:119–137. doi: 10.1007/978-1-61779-210-6_5. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. Journal of Morphology. 1951;88:231–272. [PubMed] [Google Scholar]
- Hochgreb-Hagele T, Bronner ME. A novel FoxD3 gene trap line reveals neural crest precursor movement and a role for FoxD3 in their specification. Dev Biol. 2013;374:1–11. doi: 10.1016/j.ydbio.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar P, Aplin AE. FOXD3 regulates migration properties and Rnd3 expression in melanoma cells. Mol Cancer Res. 2011;9:545–552. doi: 10.1158/1541-7786.MCR-10-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev Dyn. 2009;238:716–723. doi: 10.1002/dvdy.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–1479. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- LeDouarin N, Kalcheim C. The Neural Crest. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev Dyn. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development. 2011;138:641–652. doi: 10.1242/dev.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol. 2011;27:347–376. doi: 10.1146/annurev-cellbio-092910-154036. [DOI] [PubMed] [Google Scholar]
- Nitzan E, Krispin S, Pfaltzgraff ER, Klar A, Labosky PA, Kalcheim C. A dynamic code of dorsal neural tube genes regulates the segregation between neurogenic and melanogenic neural crest cells. Development. 2013 doi: 10.1242/dev.093294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Takeichi M. Evolution: structural and functional diversity of cadherin at the adherens junction. J Cell Biol. 2011;193:1137–1146. doi: 10.1083/jcb.201008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad MS, Sauka-Spengler T, LaBonne C. Induction of the neural crest state: control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev Biol. 2012;366:10–21. doi: 10.1016/j.ydbio.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Mizuseki K, Sasai Y. Requirement of FoxD3-class signaling for neural crest determination in Xenopus. Development. 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- Schiffmacher AT, Padmanabhan R, Jhingory S, Taneyhill LA. Cadherin-6B is proteolytically processed during epithelial-to-mesenchymal transitions of the cranial neural crest. Mol Biol Cell. 2014;25:41–54. doi: 10.1091/mbc.E13-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Bronner-Fraser M. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol Biol Cell. 2005;16:5283–5293. doi: 10.1091/mbc.E05-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KM, Labonne C. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev Cell. 2005;9:593–603. doi: 10.1016/j.devcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–1858. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Slug stability is dynamically regulated during neural crest development by the F-box protein Ppa. Development. 2006;133:3359–3370. doi: 10.1242/dev.02504. [DOI] [PubMed] [Google Scholar]
- Wang WD, Melville DB, Montero-Balaguer M, Hatzopoulos AK, Knapik EW. Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev Biol. 2011;360:173–185. doi: 10.1016/j.ydbio.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. In Situ Hybridization: A practical Approach. Oxford: Oxford University Press; 1992. Whole mount in situ hybridization of vertebrate embryos; pp. 75–83. [Google Scholar]