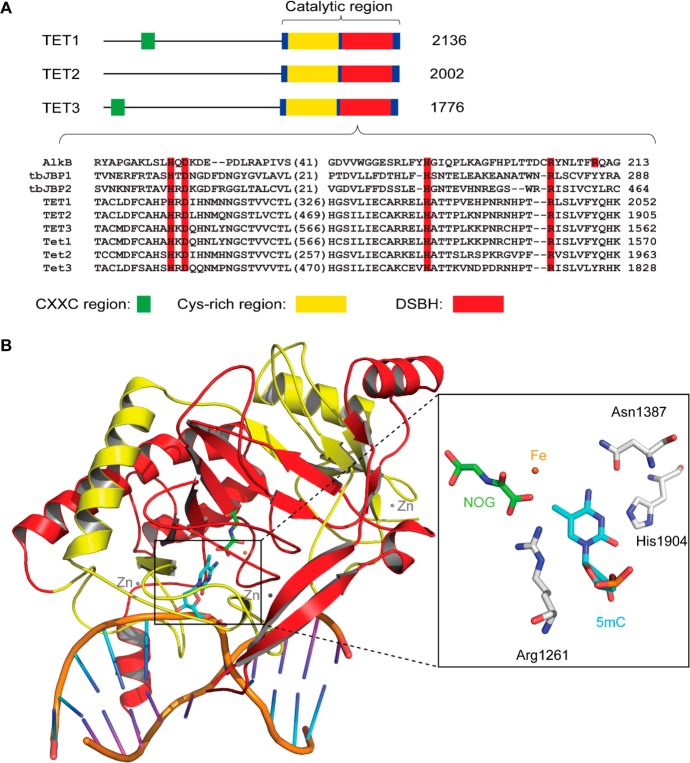
Figure 13.
Domain architecture of TET proteins. (A) TET proteins contain a DNA-binding CXXC region in the N-terminus and a catalytic core in the C-terminus. The catalytic core is composed of a Cys-rich region and a DSBH fold. The number of amino acids for each protein is indicated. Sequence alignment of the catalytic motif is shown. Sequences used in the alignment include AlkB, Trypanosoma brucei JBP1 (tbJBP1), tbJBP2, human TET1–TET3, and mouse Tet1–Tet3. Conserved iron(II)- and α-KG-binding sites are highlighted in red columns. (B) Crystal structure of the human TET2 bound to a 5mC-containing dsDNA (PDB ID 4NM6). The Cys-rich region (residues 1129–1312) and DSBH core (residues 1313–1936) are colored in yellow and red, respectively. The active-site iron is shown in orange, the α-KG analogue of NOG in green, structural zinc ions in gray, protein residues in white, flipped 5mC in blue, DNA backbone in beige, bases in the 5mC-containing DNA strand in cyan, and bases in the complementary strand in purple.