Abstract
Aim:
To investigate the effects of small interfering RNA (siRNA) knockdown of forkhead box M1 (FoxM1) on the proliferation and invasion capacities of human hepatocellular carcinoma MHCC-97H cells in vitro.
Methods:
The expression levels of FoxM1 in human hepatocellular carcinoma samples, adjacent non-hepatocellular carcinoma liver samples and MHCC-97 cell lines were detected by RT-PCR and Western blotting. FoxM1 siRNA was transfected into MHCC-97H cells with Lipofectamine 2000. Cell growth was evaluated by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, and cell cycle analysis was performed by flow cytometry. Protein expression levels were evaluated by Western blotting. Anchorage-independent growth and the invasive potency of MHCC-97H cells were measured by soft agar colony formation and a transwell cell invasion assay, respectively.
Results:
FoxM1 was over-expressed in hepatocellular carcinoma samples compared to adjacent non-hepatocellular carcinoma liver samples. FoxM1 siRNA was successfully transfected into MHCC-97H cells, resulting in the significant inhibition of FoxM1 mRNA and protein expression. Down-regulation of FoxM1 inhibited cell proliferation, caused cell cycle arrest, and decreased invasion of MHCC-97H cells. Compared with control and mock groups, the FoxM1 siRNA transfected cells showed decreased protein expressions of cyclin B1 and cyclin D1, whereas p27 protein expression was increased. Down-regulation of FoxM1 reduced the expression of matrix metalloproteinase-2 (MMP-2) and urokinase plasminogen activator (uPA).
Conclusion:
FoxM1 is functionally involved in hepatocellular carcinoma cell proliferation and invasion and is a potential target for hepatocellular carcinoma therapy.
Keywords: cell cycle, Forkhead box M1, invasion, small interfering RNA, cell cycle, proliferation, matrix metalloproteinases, urokinase plasminogen activator
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the third most common cause of cancer mortality, causing about 500 000 deaths per annum. Most HCC cases occur in either Eastern Asia, especially in China, or sub-Saharan Africa1. Currently, the prognosis for HCC is still poor, as no effective therapy has been developed2. Gene targeting is a relatively new approach to cancer therapy, and a key requirement of gene targeting therapy is to have specific and efficient targets. Thus, discovery of a new specific and efficient molecular target for HCC could be essential for reducing the mortality of this devastating malignancy.
Forkhead box M1 (FoxM1) belongs to the forkhead box transcription factor family, which is characterized by the forkhead box domain3. FoxM1 is a proliferation-associated transcription factor that has important roles in cell proliferation, organogenesis, aging and cancer4. One previous study showed that overexpression of FoxM1 is associated with the development and progression of numerous cancers5.
In this study, we examined the expression levels of FoxM1 in human HCC and adjacent non-HCC liver samples. Additionally, we studied the biological roles of FoxM1 in HCC by modulating FoxM1 expression levels in an invasive HCC cell line, MHCC-97H, using small interfering RNA (siRNA).
Materials and methods
Reagents and antibodies
Rabbit polyclonal antibodies to FoxM1, MMP-2, cyclin B1, cyclin D1, uPA, and p27 and a mouse monoclonal antibody to p21 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The rabbit polyclonal antibody to GADPH, goat anti-mouse immunoglobulin G (IgG)/horseradish peroxidase (HRP) and goat anti-rabbit IgG/HRP were from Zhongshan Biotechnology Co (Beijing, China). The protease inhibitor cocktail, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and all other chemicals were obtained from Sigma.
Patients, specimens, and cell lines
Thirty-two pairs of human HCC samples and adjacent non-HCC liver samples were obtained from patients who underwent surgical resection. All samples were immediately frozen and stored at −80 °C. Informed consent was obtained from all patients, and this study was approved by our institutional review board. The HCC cell lines, MHCC-97H, and MHCC-97L, used in this study were obtained from the Liver Cancer Institute, Fudan University (Shanghai, China), and maintained in DMEM containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37 °C with 5% CO2.
Transfection of siRNA
RNA interference mediated by duplexes of 21-nucleotide RNA was performed in MHCC-97H cells. FoxM1 siRNA (GGACCACUUUCCCUACUUU) and mock siRNA (GGACCUGUAUGCGUACAUU) were synthesized by Shanghai GenePharma Co (Shanghai, China). Negative control siRNA duplexes (siRNA-NC), labeled with the fluorophore FAM, was used to detect the transfection efficiency. The siRNA-NC did not target any known mammalian gene and was synthesized by Shanghai GenePharma Co (Shanghai, China). Transfection of siRNA was carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the procedure recommended by the manufacturer. Six hours after transfection with siRNA-NC at various concentrations, the cells were analyzed on a FACS Calibur flow cytometer equipped with Cell Quest software (Becton Dickinson, San Jose, CA, USA).
Reverse transcription polymerase chain reaction (RT-PCR) analysis
Total RNA was isolated from the human samples and different cell groups using the RNAfast200 Total RNA Extract Kit (Fastgene, Shanghai, China), and 2 μg RNA was reverse transcribed to cDNA by the RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, MBI, Lithuania). Two pairs of primers for the FoxM1 gene were used in this study. One pair of primers was 5′-AACCGCTACTTGACATTGG-3′ (forward) and 5′-GCAGTGGCTTCATCTTCC-3′ (reverse). The other primer pair was 5′-CACCCCAGTGCCAACCGCTACTTG-3′(forward) and 5′-AAAGAGGAGCTATCCCCTCCTCAG-3′ (reverse). Primers for the GAPDH gene were as follows: 5′-GGTATCGTGGAAGGACCATGAC-3′ (forward) and 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ (reverse). The PCR was performed in a 25 μL volume reaction buffer. After a 5 min pretreatment at 94 °C, 30 PCR cycles of denaturing at 94 °C for 30 s, annealing at 58 °C for 45 s, and extension at 72 °C for 45 s were performed. The PCR products were resolved on a 2% agarose gel.
Western blotting
Cells and human samples were lysed in ice-cold RIPA lysis buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mmol/L NaCl and 10 mmol/L Tris-HCl) containing a protease inhibitor cocktail. The total protein concentration was determined using a Bio-Rad protein assay reagent (Bio-Rad, Hercules, CA, USA). Equivalent amounts of proteins (30 μg) were then separated by 12% SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA). After being blocked in Tris buffered saline (TBS) containing 5% non-fat milk, the membranes were incubated with primary antibodies at 4 °C for 12 h and then with horseradish peroxidase (HRP) conjugated anti-goat or anti-rabbit antibody (Zhongshan, Beijing, China) at a dilution of 1:3000 at room temperature for 1 h. Signals were detected on X-ray film using the ECL detection system (Pierce, Rockford, IL, USA). Equal protein loading was assessed by the expression of GAPDH.
Cell proliferation assay
Cell viability was measured by MTT assay. MHCC-97H cells were seeded at 5×103 per well in 96-well flat-bottom plates one day before transfection. The MTT assay was performed just before transfection as well as 24, 48, and 72 h after transfection. For the assay, 20 μL of 5 mg/mL MTT was added to each well, then the cells were incubated for 4 h before 180 μL DMSO was added. After the insoluble crystals were completely dissolved, the absorbance values at 570 nm were measured using a microplate reader (Bio-Rad, Hercules, CA, USA).
Cell cycle analysis
Cell cycle progression was determined by flow cytometry of propidium iodide (PI) stained cells on a FACS Calibur flow cytometer (Becton Dickinson, San Jose, CA, USA). Forty-eight hours after transfection, cells were trypsinized, collected and washed twice with PBS. Then the cells were fixed with cold 70% ethanol for at least 1 h at 4 °C. Before flow cytometric analysis, the cells were incubated with RNase at 0.01 mg/mL and PI at 50 μg/mL in the dark at room temperature for 30 min. A total of 104 cells were analyzed for each sample. Cell cycle analysis was performed by ModFit LT version 3.0 software (Verity Software House, Topsham, ME, USA). Experiments were done in triplicate.
Soft agar colony formation assay
MHCC-97H cells were transfected with the FoxM1 siRNA (100 nmol/L) or mock siRNA (100 nmol/L). Twenty-four hours after transfection, cells were trypsinized, and 5×103 cells were mixed with a 0.3% agar solution in DMEM containing 10% FBS and layered on top of a 0.6% agar layer in six-well tissue culture plates. The plates were incubated for 2–3 weeks at 37 °C in a 5% CO2 atmosphere until colonies were formed. Only colonies containing more than 50 cells were counted. The results were reported as the mean number of colonies observed in five randomly chosen microscope fields. Experiments were done in triplicate.
Cell invasion assay
The invasive behaviors of the MHCC-97H cells were tested using a Matrigel transmembrane invasion assay. Transwell chambers (Millipore, Bedford, MA, USA) (8 μm pore size) were coated with Matrigel (15 μg/filter). Cells (2.0×104) in serum-free medium were plated into the upper chamber and bottom wells were filled with complete medium. Cells were allowed to invade across the Matrigel-coated membrane for 48 h at 37 °C in 5% CO2. After incubation, cells were removed from the upper surface of the filter by scraping with a cotton swab. The invaded cells that adhered to the bottom of the membrane were fixed with methanol and stained with a crystal violet solution. The number of cells that penetrated the membrane was determined by counting the mean cell number of five randomly selected high-power fields. Experiments were done in triplicate.
Statistical analysis
Results are expressed as mean±standard deviation (SD). Statistical evaluation of the data was performed with one-way ANOVA. Pair-wise comparisons were conducted by a Student Newman-Keuls test. Two-tailed P<0.05 was considered statistically significant. All analyses were performed using SPSS software version 13.0 (SPSS Inc, Chicago, IL, USA).
Results
Expression of FoxM1 in HCC and adjacent non-HCC liver samples
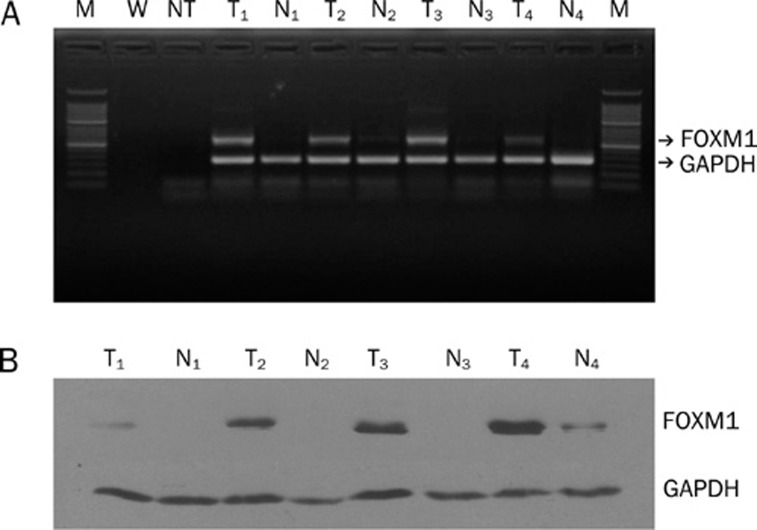
Using RT-PCR, FoxM1 mRNA was found to be significantly overexpressed in 87.5% of HCC specimens, whereas it was detected in only 12.5% of adjacent non-HCC liver samples (P<0.05) (Figure 1A). To further confirm these data, we evaluated the protein expression of FoxM1 in these samples by Western blotting. The results showed that FoxM1 protein expression could be detected in 93.75% of HCC samples but only 12.5% of adjacent non-HCC liver samples (P<0.05). Similar to the gene expression pattern, FoxM1 was obviously increased in HCC as compared with adjacent non-HCC liver samples (Figure 1B).
Figure 1.
FoxM1 expression in tumor and adjacent non-tumor liver samples detected by RT-PCR (A) and Western blotting (B). M: 50 bp marker; W: water; NT: no template mixture; T: tumor tissue; N: corresponding non-tumor tissue.
Expression of FoxM1 in MHCC-97 cell lines
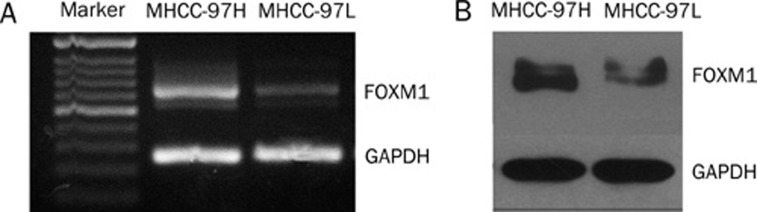
The baseline mRNA levels of FoxM1 in MHCC-97 cell lines6, 7, including MHCC-97H (an invasive HCC cell line) and MHCC-97L (a less invasive HCC cell line), were determined by RT-PCR. The results showed that the FoxM1 gene was highly expressed in MHCC-97H compared with MHCC-97L (P<0.05) (Figure 2A). Consistent with the gene expression data, the relative protein expression of FoxM1 was higher in MHCC-97H compared with MHCC-97L, as determined by Western blotting (P<0.05) (Figure 2B). The results indicated that the expression of FoxM1 may coincide with malignant phenotypes in HCC.
Figure 2.
Expression of FoxM1 in MHCC-97 cell lines detected by RT-PCR (A) and Western blotting (B).
Transfection efficiency analysis
To estimate the siRNA oligonucleotide transfection efficiency in MHCC-97H cells, flow cytometry was performed six hours after transfection with FAM-labeled siRNA-NC. We found that efficiency increased in a dose-dependent manner using siRNA concentrations of 25 to 100 nmol/L and achieved a satisfactory transfection efficiency of 90.67%±2.34% using 100 nmol/L siRNA. Thus, the concentration of 100 nmol/L siRNA oligonucleotides was selected as an optimal concentration in the following experiments.
siRNA silencing of FoxM1 at the mRNA and protein levels
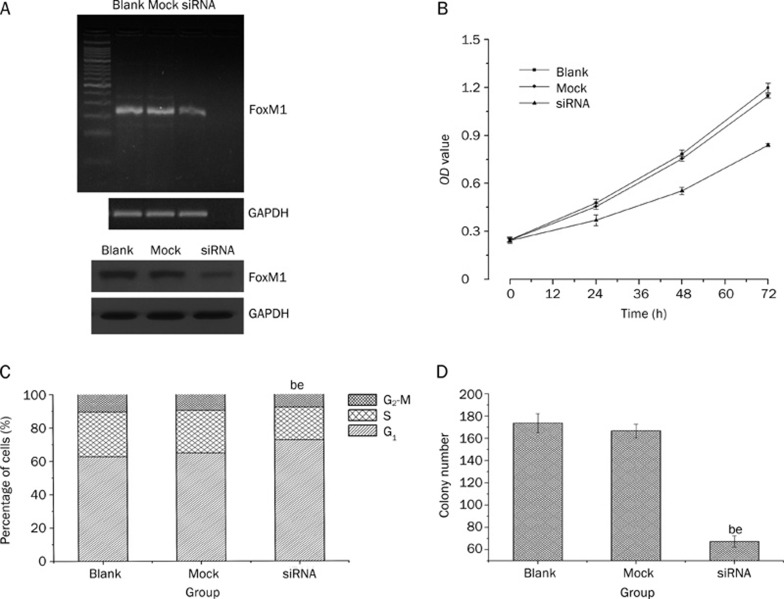
MHCC-97H cells, which have higher levels of FoxM1 expression, were transiently transfected with 100 nmol/L FoxM1 or mock siRNA. Total RNA and protein were isolated and analyzed by RT-PCR and Western blotting at 48 h and 72 h after transfection. Compared with the blank (no siRNA) and mock transfected cells, the expression of FoxM1 was obviously suppressed in cells transfected with FoxM1 siRNA at both the mRNA and protein levels (P<0.05) (Figure 3A).
Figure 3.
Down-regulation of FoxM1 inhibited cell proliferation and induced cell cycle arrest (S phase) in MHCC-97H cell line. Successful knockdown of FoxM1 shown by RT-PCR and Western blotting (A); Down-regulation of FoxM1 inhibited cell proliferation (B); Down-regulation of FoxM1 induced S phase arrest (C); Down-regulation of FoxM1 inhibited colony formation (D). bP<0.05 vs Blank group. eP<0.05 vs Mock group.
Down-regulation of FoxM1 expression by siRNA inhibited cell growth and arrested cell cycle at S-phase
As shown in Figure 3B, down-regulation of FoxM1 expression in MHCC-97H cells caused significant inhibition of cell proliferation compared with the blank and mock transfected cells. (P<0.05). Flow cytometry was used to assess whether down-regulation of FoxM1 expression in MHCC-97H cells was associated with alterations in cell cycle progression. As shown in Figure 3C, the percentage of MHCC-97H cells in S phase in FoxM1 siRNA-transfected cells was decreased relative to the blank and mock transfected cells (P<0.05). These results indicated that down-regulation of FoxM1 expression suppressed cell cycle progression in MHCC-97H cells.
Down-regulation of FoxM1 expression inhibited colony formation
Next, we determined whether knocking down FoxM1 expression affected the anchorage-independent growth of MHCC-97H cells. As shown in Figure 3D, down-regulation of FoxM1 in MHCC-97H cells inhibited anchorage-independent growth as evidenced by a decrease in the number of colonies on soft agar in the FoxM1 siRNA transfected cells compared to the blank group and mock transfected cells (P<0.05).
Down-regulation of FoxM1 expression suppressed cell invasion
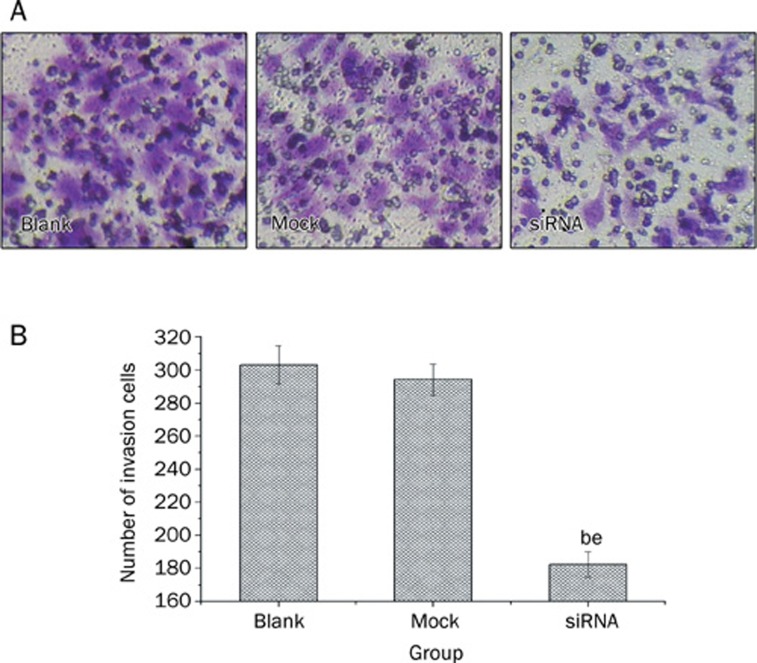
To determine whether down-regulation of FoxM1 affected the invasive ability of MHCC-97H cells, we performed cell invasion assays using a transwell system. We demonstrated that down-regulation of FoxM1 by FoxM1 siRNA inhibited the invasiveness of MHCC-97H cells compared with blank and mock transfected cells, as indicated by a marked decrease in the number of cells that invaded the bottom well (P<0.05, Figure 4). These results suggest that FoxM1 promotes cell invasion in MHCC-97H cells.
Figure 4.
Down-regulation of FoxM1 suppressed cell invasion in the MHCC-97H cell line as measured by a transwell cell invasion assay. bP<0.05 vs Blank group. eP<0.05 vs Mock group.
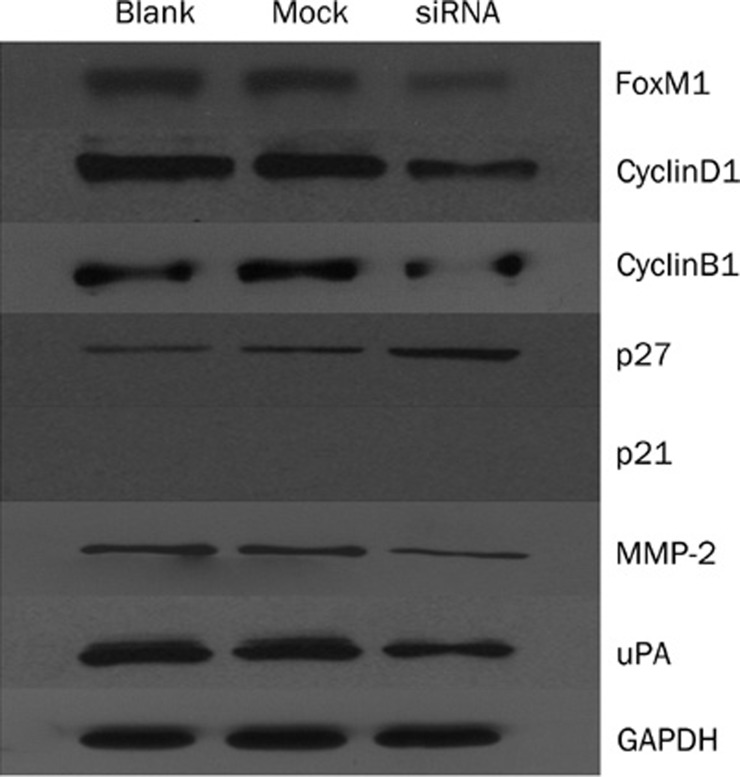
Down-regulation of FoxM1 expression altered the expression of cell cycle and invasion related proteins
As shown in Figure 5, down-regulation of FoxM1 expression in MHCC-97H cells decreased the expression of cyclin B1 and cyclin D1 and elevated the expression of p27 compared with blank and mock transfected cells (P<0.05). At the same time, down-regulation of FoxM1 expression in MHCC-97H cells reduced the expression of MMP-2 and uPA in the FoxM1 siRNA transfected cells compared with the blank and mock transfected cells (P<0.05).
Figure 5.
Down-regulation of FoxM1 altered the expression of cell cycle and invasion related proteins.
Discussion
The forkhead box family is an extensive family of transcription factors, consisting of more than 50 mammalian proteins. Members of this family share homology in the winged helix DNA-binding domain and are important for a wide spectrum of biological processes, including metabolism, development, differentiation, proliferation, apoptosis, migration, invasion and longevity3. FoxM1, one member of this family, is a critical regulator of cell cycle progression and has important roles in cell proliferation, organogenesis, aging and cancer4. In vitro, loss of FoxM1 is associated with cell cycle arrest and leads to lesion of mitotic spindle integrity. In vivo, loss of FoxM1 is embryonic lethal due to a failure to enter mitosis8, 9, 10. Recent studies showed that FoxM1 was overexpressed in several kinds of cancer, such as lung cancer, glioblastomas and gastric cancer, and had an important role in the development and progression of those cancers5, 11, 12, 13.
FoxM1 expression is tightly associated with proliferation and is extinguished in differentiated or resting cells that have exited the cell cycle. In normal tissues, the expression of FoxM1 is restricted to embryonic tissues and a few adult endocrine glands14. FoxM1 is strongly expressed in mouse fetal liver but not in the adult liver. However, FoxM1 is highly expressed in HCC and HCC cell lines. The data above indicate that FoxM1 is a potential specific target for HCC therapy. Interestingly, a recent study showed that lower expression of FoxM1 in HCC was associated with prolonged disease-free survival after curative liver resection and validated FoxM1 as a prognostic marker for HCC15.
In this study, we first determined the expression level of FoxM1 in HCC and adjacent non-HCC liver samples and found that FoxM1 was more highly expressed in the HCC samples. To elucidate the functional relevance of FoxM1 in HCC, we modulated the FoxM1 expression level in an invasive HCC cell line, MHCC-97H, with siRNA. Our results demonstrated that down-regulation of FoxM1 expression in MHCC-97H cells by siRNA could inhibit cell proliferation, induce cell cycle arrest and inhibit cell invasion, indicating that FoxM1 is a potential therapeutic target for the treatment of HCC.
Abnormal cell proliferation is an important characteristic of malignant tumors, including HCC, which suggests that cell cycle arrest could be an effective method of therapy for malignant tumors. In this study, we found that down-regulation of FoxM1 expression by siRNA could inhibit cell proliferation. Furthermore, cell cycle analysis showed that the percentage of G0-G1 phase cells was significantly elevated in the FoxM1 siRNA transfected cells compared with blank and mock transfected cells, indicating an S-phase arrest of the cell cycle. Cell proliferation is regulated by cell cycle machinery and depends on the balance between regulators of cyclin-dependent kinases, specifically positive factors (cyclins) and CDK inhibitors (CDKI)16. In the present study, we detected the expression of cyclins (cyclinB1 and cyclinD1) and CDKI (p21 and p27). Our results showed that down-regulation of FoxM1 expression by siRNA caused a marked reduction in cyclin B1 and cyclin D1 and elevated the expression of p27. However, we did not detect the expression of p21 in this cell line. We propose that like C33A, a cervical cancer cell line, p21 may be absent in this cell line17. These results suggest that FoxM1 affects the cell cycle of MHCC-97H cells by regulating the expression levels of some cyclins (cyclin D1 and cyclin B1) and CDKI (p27). In addition to the above mechanisms, a recent study suggests that cellular senescence caused by FoxM1 depletion may be involved in the inhibition of cell proliferation, suggesting yet another mechanistic target for cancer therapy11.
Acquisition of cell migratory and invasive abilities is essential for cancer invasion and metastasis. One of the key steps in the cancer invasion and metastatic cascade involves the disruption of extracellular matrix (ECM) and basement membranes, which permits tumor cells to access a distant site. A group of proteins called proteolytic enzymes were shown to be involved in cancer invasion and metastasis18. The key proteases are the serine proteases (plasmins), the urokinase plasminogen activator (uPA), cysteine proteases and matrix metalloproteinases (MMPs). Among these enzymes, MMPs and uPA have important roles in cancer cell invasion and metastasis19, 20. In the present study, we found that down-regulation of FoxM1 expression by siRNA decreased the invasive ability of MHCC-97H cells. When we detected the expression of MMP2, a well-studied and critical member of the MMP family, and uPA, our results showed that down-regulation of FoxM1 expression by siRNA inhibited the expression of MMP-2 and uPA. It is therefore highly likely that the abolished HCC invasion due to inhibition of FoxM1 may primarily be induced by inhibition of MMP-2 and uPA.
In conclusion, our results showed that down-regulation of the FoxM1 expression level in MHCC-97H cells by FoxM1-specific siRNA decreased cell proliferation and invasion ability. Furthermore, decreasing the FoxM1 expression level resulted in down-regulation of cyclins B1 and D1 and up-regulation of p27. Down-regulation of FoxM1 also reduced the expression of MMP-2 and uPA. This study provides clear evidence that FoxM1 is functionally important in the development and progression of HCC and may serve as a new target for HCC therapy.
Author contribution
Chang LIU and Yi LÜ designed research; Qi-fei WU, Dong LIU, Lei LEI, Min TIAN, and Rui-tao WANG performed research; Ming-hui TAI analyzed data; Qi-fei WU wrote the paper.
Acknowledgments
We are grateful to Prof Xin-yang WANG for his technical assistance. We also thank Dr Lei LI and Lin-lin ZHANG for their valuable help.
This project was supported by National Natural Science Foundation of China (No 30901447/C160402).
References
- Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Cao LQ, Chen XL, Wang Q, Huang XH, Zhen MC, Zhang LJ, et al. Upregulation of PTEN involved in rosiglitazone-induced apoptosis in human hepatocellular carcinoma cells. Acta Pharmacol Sin. 2007;28:879–87. doi: 10.1111/j.1745-7254.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–61. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye QH, et al. A decade's studies on metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:187–96. doi: 10.1007/s00432-003-0511-1. [DOI] [PubMed] [Google Scholar]
- Tian J, Tang ZY, Ye SL, Liu YK, Lin ZY, Chen J, et al. New human hepatocellular carcinoma (HCC) cell line with highly metastatic potential (MHCC97) and its expressions of the factors associated with metastasis. Br J Cancer. 1999;81:814–21. doi: 10.1038/sj.bjc.6690769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, Dennewitz MB, et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- Zeng J, Wang L, Li Q, Li W, Björkholm M, Jia J, et al. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27kip1. J Pathol. 2009;218:419–27. doi: 10.1002/path.2530. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–9. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–9. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, Pinna F, Ladu S, Pellegrino R, Simile MM, Frau M, et al. Forkhead box M1B is a determinant of rat susceptibility to hepatocarcinogenesis and sustains ERK activity in human HCC. Gut. 2009;58:679–87. doi: 10.1136/gut.2008.152652. [DOI] [PubMed] [Google Scholar]
- Wakino S, Kintscher U, Kim S, Yin F, Hsueh WA, Law RE. Peroxisome proliferator-activated receptor gamma ligands inhibit retinoblastoma phosphorylation and G1→S transition in vascular smooth muscle cells. J Biol Chem. 2000;275:22435–41. doi: 10.1074/jbc.M910452199. [DOI] [PubMed] [Google Scholar]
- Chan DW, Yu SY, Chiu PM, Yao KM, Liu VW, Cheung AN, et al. Over-expression of FOXM1 transcription factor is associated with cervical cancer progression and pathogenesis. J Pathol. 2008;215:245–52. doi: 10.1002/path.2355. [DOI] [PubMed] [Google Scholar]
- Duffy MJ. The role of proteolytic enzymes in cancer invasion and metastasis. Clin Exp Metastasis. 1992;10:145–55. doi: 10.1007/BF00132746. [DOI] [PubMed] [Google Scholar]
- Rabbani SA, Mazar AP. The role of the plasminogen activation system in angiogenesis and metastasis. Surg Oncol Clin N Am. 2001;10:393–415. [PubMed] [Google Scholar]
- Cox G, O'Byrne KJ. Matrix metalloproteinases and cancer. Anticancer Res. 2001;21:4207–19. [PubMed] [Google Scholar]