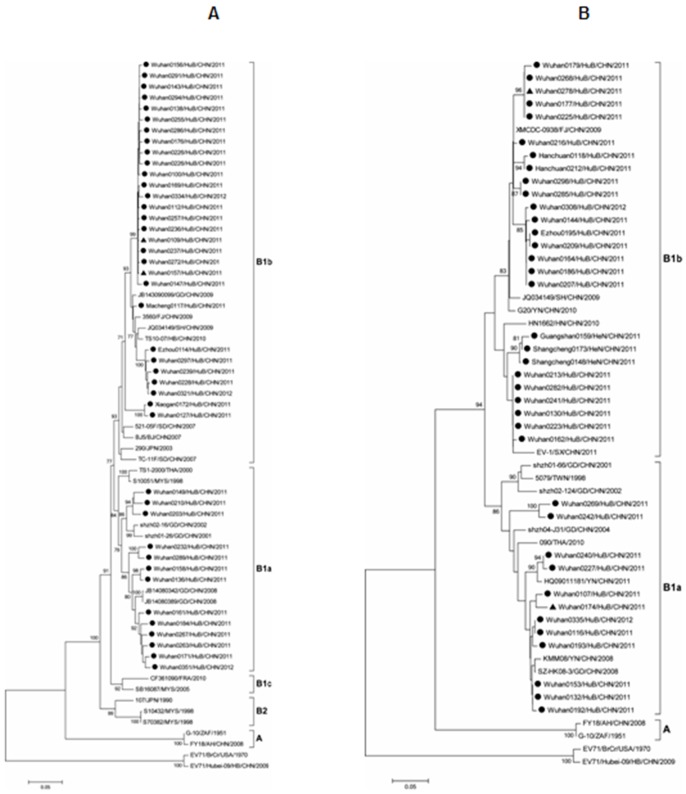
Figure 7. Molecular characterization and phylogenetic analyses of coxsackievirus A16 (CVA16) isolates.
(A) Phylogenetic dendrogram constructed by the neighbor-joining method based on 891-bp complete VP1 sequence of the 42 CVA16 strains isolated in this study. (B) Phylogenetic dendrogram constructed by the neighbor-joining method based on 209-bp partial VP3-VP1 sequence of the 38 CVA16 strains. No VP3-VP1 sequences from B1c and B2 reference strains were available from GenBank. Strains indicated by a filled circle (•) and triangle (▴) are CVA16strains isolated from mild cases and severe cases, respectively, during the HFMD outbreak. Bootstrap values with >70 replications are shown at the branch nodes as percentages. The scale bar represents the genetic distance. Strain enterovirus 71 BrCr and another enterovirus 71 Hubei-09 strain from China were used as out groups.