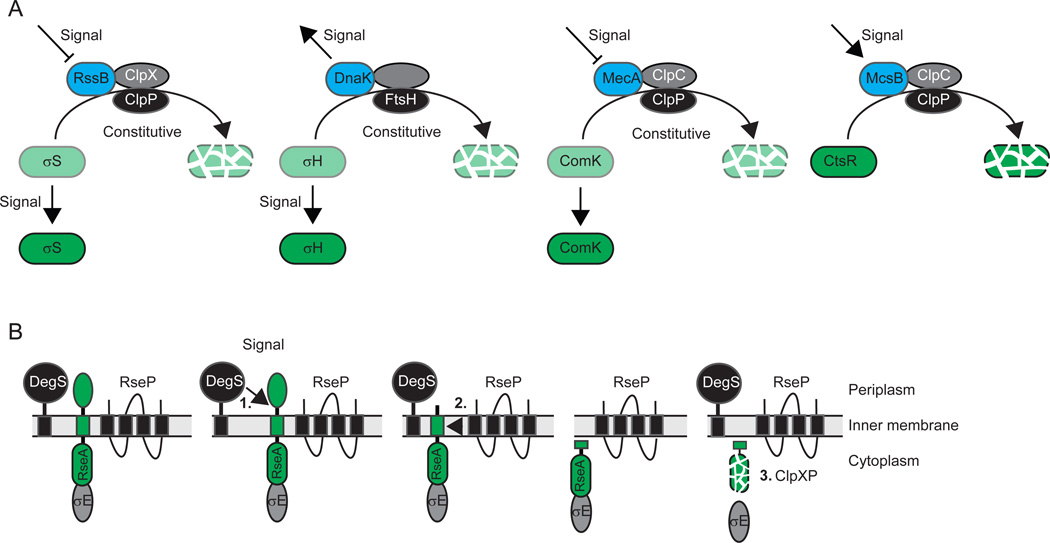
Figure 2. Principles of regulated proteolysis from stress response studies.
(A) Representative examples of circuit design for the regulated destruction of regulatory cytoplasmic proteins. In all four examples, the level of green indicates the accumulation level of the substrate. Adaptors are indicated in blue, proteases in black and AAA+ proteins in grey. Note that in FtsH, the protease and AAA+ domains are part of the same polypeptide.
(B) Representative example for the regulated destruction of a regulatory inner membrane protein by RIP. The bitopic inner membrane protein substrate RseA is indicated in green. Cleavage by DegS, the site-1-protease, is immediately followed by cleavage by RseP, the site-2-protease, and degradation by ClpXP.