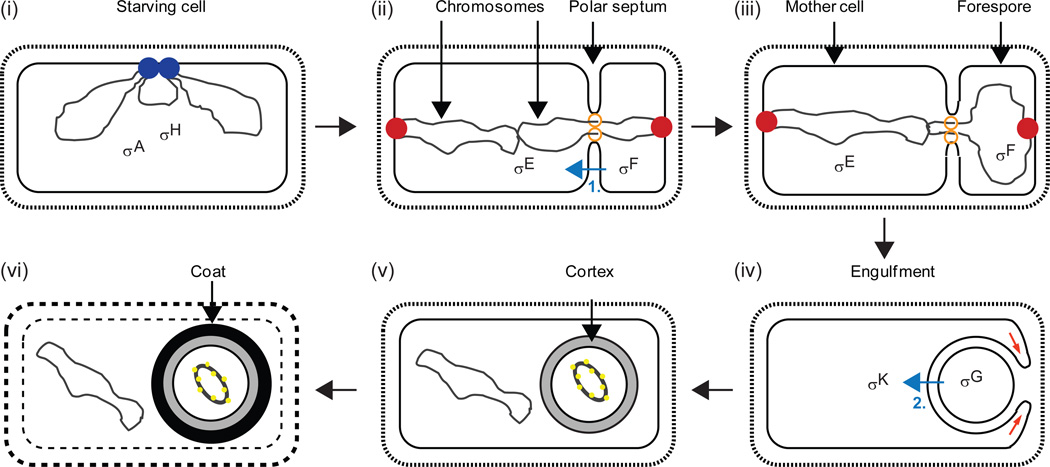
Figure 4. Morphological changes and sigma factors during B. subtilis endospore formation.
(i) The DNA replication machinery (blue) completes chromosome (black) duplication in a starving cell. The major vegetative sigma factor, σA, and the alternative sigma factor, σH, begin to direct transcription of sporulation genes. (ii) A region near each chromosome’s origin of replication is bound by a complex of proteins (red) attached to the membrane at opposite cell poles. The polar septum forms with a DNA translocase (orange) at the annulus. About one-third of the origin-proximal region of one chromosome is located in the smaller forespore compartment, creating genetic asymmetry for about 15 min. σF becomes active in the forespore and initiates a signaling pathway (blue arrow labelled “1”) that leads to proteolytic activation of σE in the mother cell as described in Figure 5. (iii) Most of one chromosome is translocated into the forespore. (iv) The mother cell membrane migrates around the forespore membrane (red arrows) as engulfment begins. σG becomes active in the forespore and initiates a signaling pathway (blue arrow labelled “2”) that leads to proteolytic activation of σK in the mother cell as described in Figures 6 and 7. (v) Cortex forms between the inner and outer forespore membranes. The forespore chromosome is condensed into a toroid by binding of small, acid-soluble spore proteins (yellow). (vi) Spore coat proteins complete their assembly on the forespore surface. The mother cell lyses. Adapted from (Kroos, 2007).