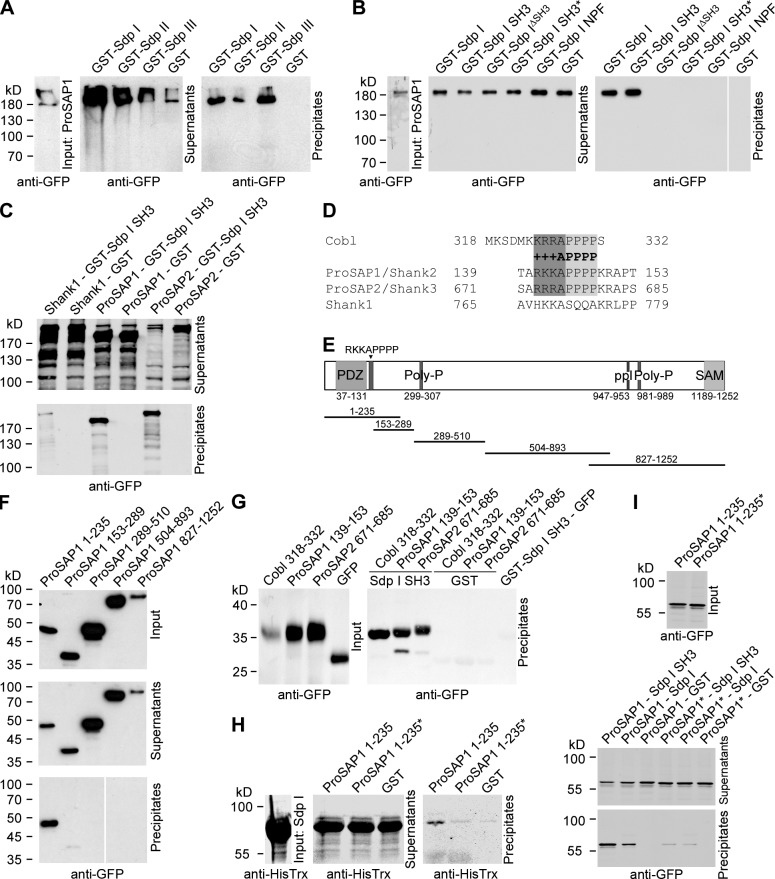
Figure 3.
Identification of ProSAP1/Shank2 and ProSAP2/Shank3 as postsynaptically enriched Syndapin I interaction partners. (A) GST–syndapin I, II, and III specifically precipitate GFP-ProSAP1 expressed in HEK293 cells. (B) Coprecipitation analysis with GST–syndapin I and deletion mutants thereof. The SH3 domain is critical and sufficient for binding. A mutant SH3 domain (P434L; SH3*) did not bind. White lines indicate lanes omitted from blots (B and F). (C) Syndapin I SH3 precipitates GFP-ProSAP1 and GFP-ProSAP2 but not GFP-Shank1. (D) Alignment of +++APPPP motifs in ProSAP1 (NCBI Protein database accession no. NP_001004133), ProSAP2 (accession no. NP_067708), and Cobl (accession no. NP_766084; conserved amino acids are highlighted) and of corresponding residues in Shank1 (accession no. Q9WV48). (E) Scheme of rat ProSAP1b and deletion mutants used. Indicated are the N-terminal PDZ domain (medium grey), several proline-rich motifs (dark grey lines), and the C-terminal SAM (sterile alpha motif) domain (light grey). (F) GST–syndapin I precipitated GFP-ProSAP1 1–235 but none of the other ProSAP1 deletion mutants. (G) GFP fusion peptides encompassing the +++APPPP motifs of ProSAP1, ProSAP2, and Cobl associated with syndapin I SH3. (H and I) RKKAPPPPKR to GAGAAAAAAG mutation (amino acids 141–150 in ProSAP1; ProSAP1 1–235*) disrupted direct binding of ProSAP1 to syndapin I in both in vitro reconstitutions with purified proteins (H) and in coprecipitation analyses (I).