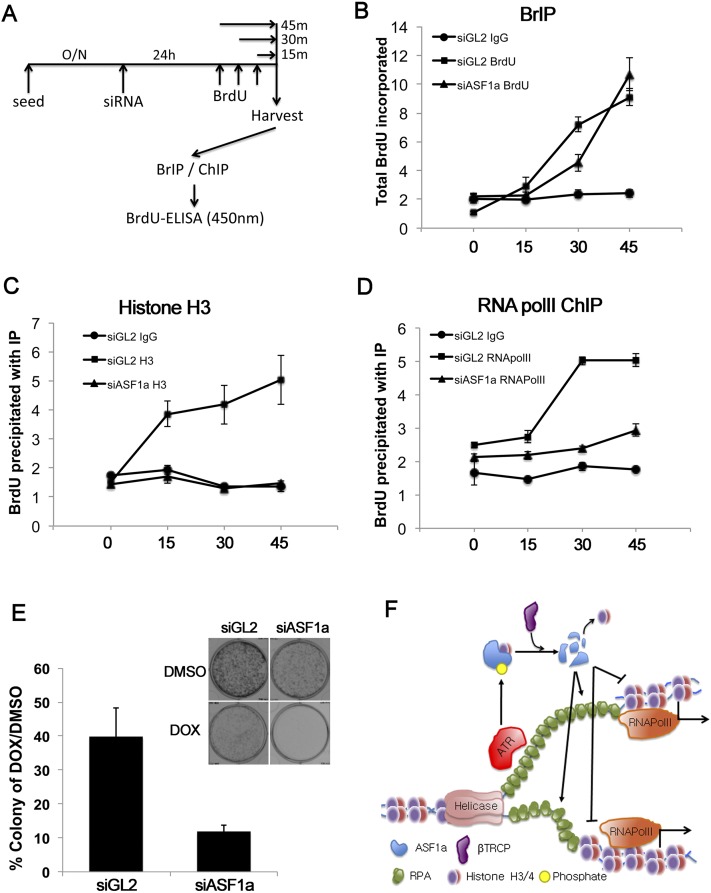
Figure 7.
Depletion of ASF1a inhibits the loading of RNA polymerase II at newly synthesized DNA. (A–D) After 24 h of depletion of ASF1a, BrdU was added during the indicated time (shown in A). BrdU-incorporated DNA was captured by BrIP (B), histone H3 ChIP (C), or RNA polymerase II (polII) ChIP (D). BrdU-incorporated DNA eluted from BrIP or ChIP was analyzed by ELISA with anti-BrdU antibody. (E) The percentage of viable colonies after exposure of HeLa cells to DOX relative to DMSO with or without depletion of ASF1a (left) and representative examples of plates with colonies (top right). Mean ± SD from three independent experiments. (F) Schematic model of transcriptional repression of genes near DOX-induced clusters of stalled replication forks. ASF1a promotes nucleosome loading on the newly replicated DNA. This chromatinization promotes RNA polymerase II loading and suppresses unwinding or nuclease activity on the newly replicated dsDNA. ASF1a is itself degraded by ATR and CRL1βTRCP-dependent pathways activated by RPA-coated ssDNA that is present at stalled forks and that increases as more ssDNA is generated after dechromatinization of the newly replicated DNA.