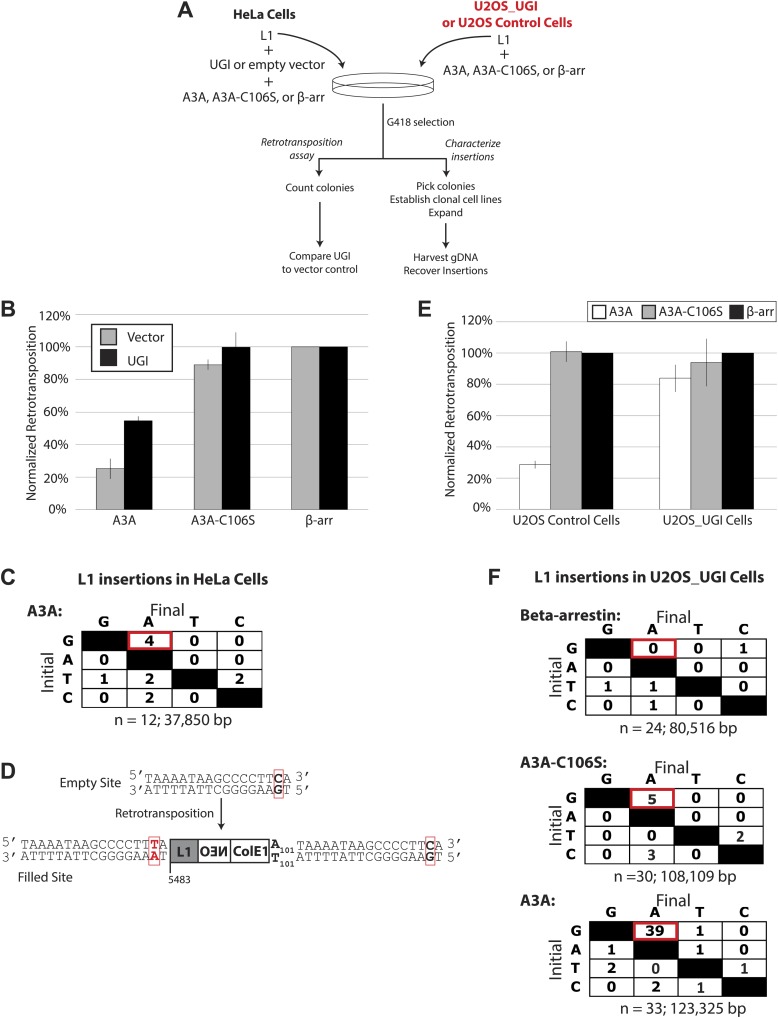
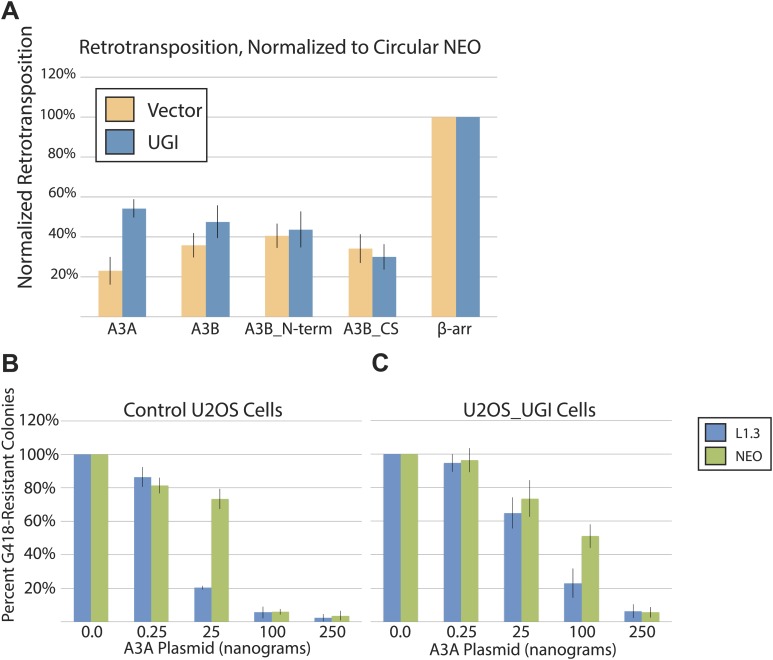
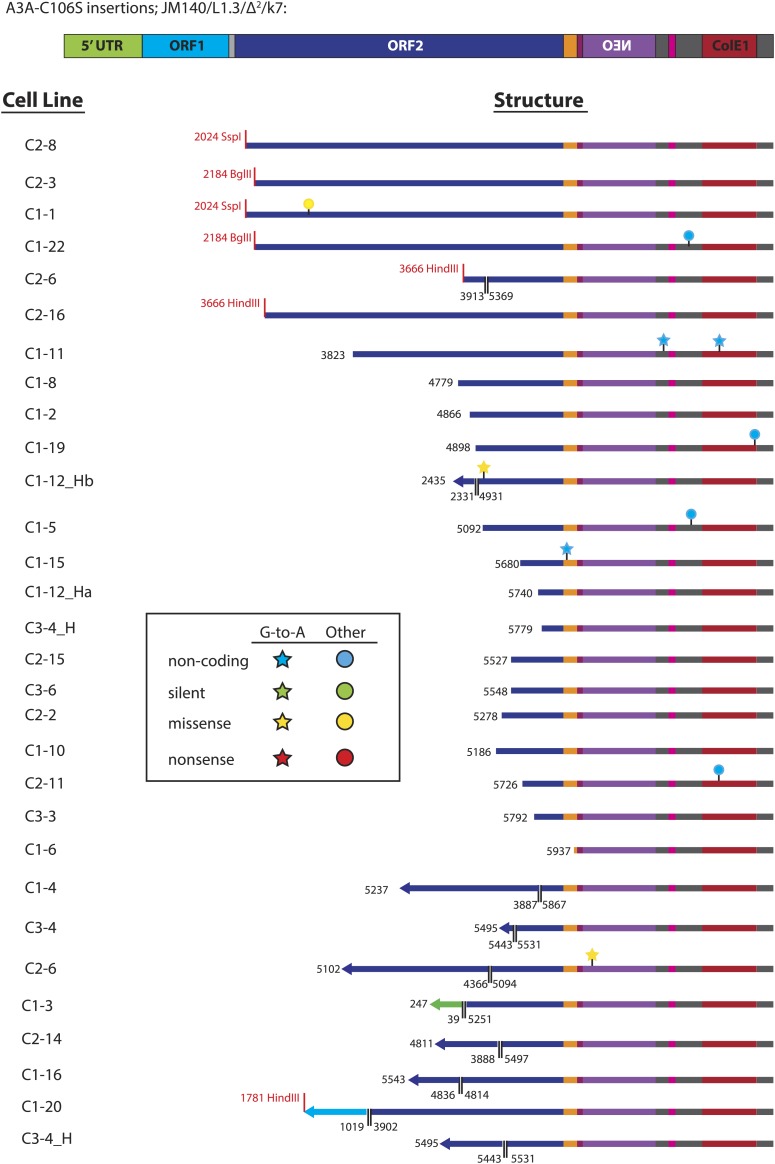
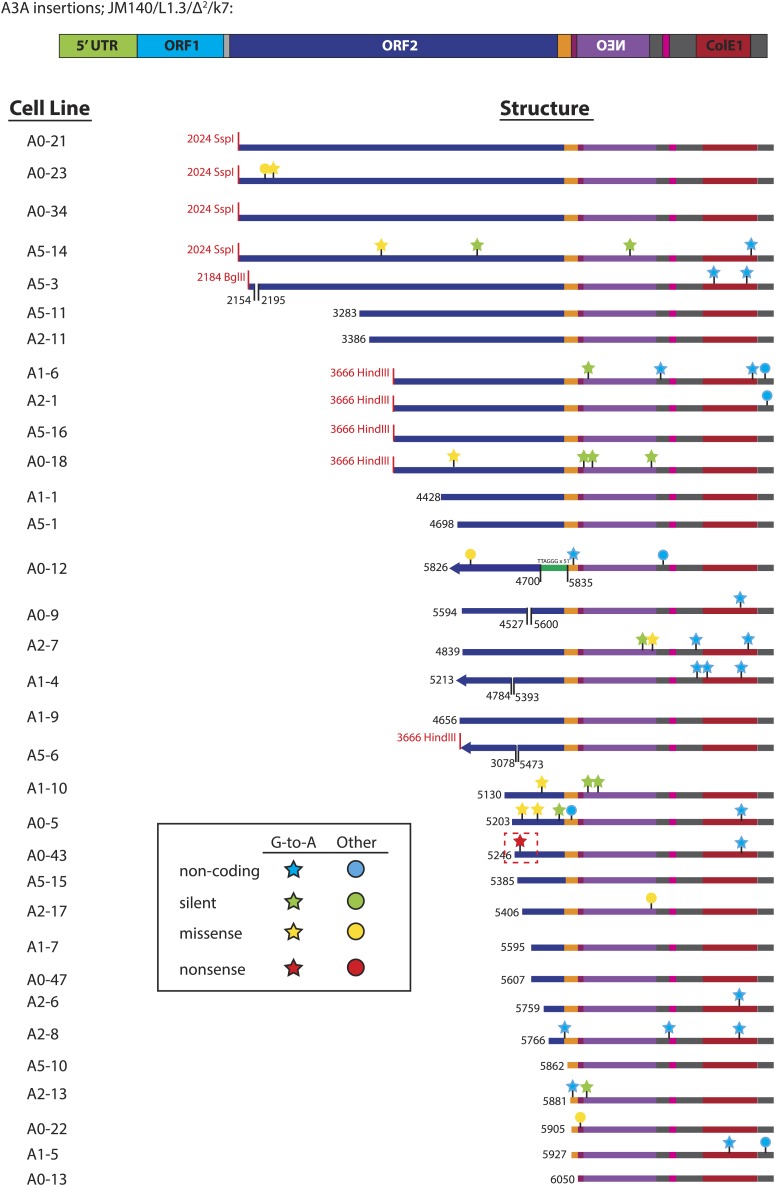
Figure 3. UGI expression alleviates A3A-mediated inhibition of L1 retrotransposition and allows detection of deaminated L1 retrotransposition events.
(A) Experimental strategy: cells (HeLa, upper left, or U2OS, upper right) were co-transfected with the indicated expression plasmids and assayed for L1 retrotransposition (bottom left branch) or insertion analysis (bottom right branch). (B) UGI expression alleviates A3A-mediated retrotransposition inhibition: the x-axis indicates the co-expression vector. The y-axis depicts the efficiency of L1 retrotransposition. Shown are results of experiments in the presence (black bars) or absence (gray bars) of UGI. The results were normalized as in Figure 1—figure supplement 1A. Data are expressed as the mean percent retrotransposition derived from three independent experiments consisting of two technical replicates each, with error bars representing the standard deviation among all six technical replicates. (C) DNA sequencing results: the left column indicates the engineered L1 sequence. The top row indicates sequence changes observed in recovered L1 insertions; n indicates the number of characterized retrotransposition events. The total retrotransposed sequence observed (in bp) includes the L1 sequence and the mneoI/ColE1 cassette. (D) An L1 insertion harboring mismatched TSDs: The pre- (empty) and post- (filled) L1 integration sites are shown. The truncation point (bp 5483) and structure of the engineered retrotransposed L1 are indicated. Deamination of single-strand genomic DNA in the pre-integration site (red rectangle) leads to inexact TSDs in the post-integration site (red lettering/rectangles). (E) Stable UGI expression alleviates A3A-mediated L1 inhibition: The x-axis indicates the U2OS cell line. The y-axis depicts the efficiency of L1 retrotransposition. Shown are the effects of wild-type A3A (white bars), A3A-C106S (gray bars), and β-arrestin (β-arr, black bars) control on L1 retrotransposition. Data were normalized to controls conducted with a circular NEO expression vector (Figure 3—figure supplement 1 B–C). Data are expressed as the mean percent retrotransposition derived from three independent experiments consisting of two technical replicates each, with error bars representing the standard deviation among all six technical replicates. (F) DNA sequencing results: The left column indicates the engineered L1 sequence. The top row indicates sequence changes observed in recovered L1 insertions; n indicates the numbers of characterized retrotransposition events. The total retrotransposed sequence observed (in bp) includes the L1 sequence and the mneoI/ColE1 cassette.