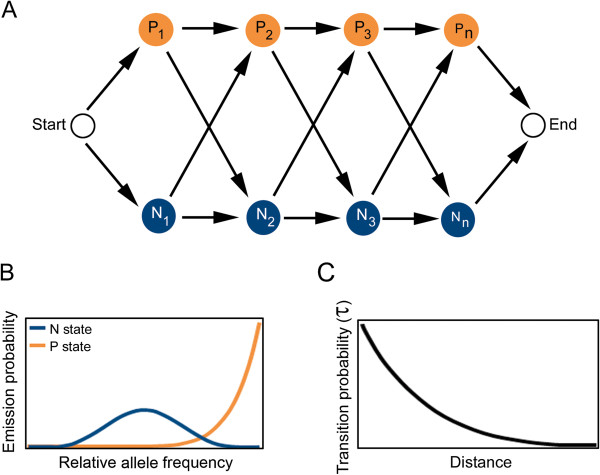
Figure 2.
Hidden Markov Model used to predict genomic regions linked to the phenotype of interest. A: each marker site is modeled to be in a neutral state (N-state, blue circles) or in a state of being linked to the phenotype of interest (P-state, orange circles) based on its observed relative variant frequency in the pool of segregants. B: emission probabilities for respectively the neutral (blue curve) and the phenotype-linked states (orange line) as a function of the relative variant frequencies, modeled by a beta-binomial distribution with respective parameters α and β. C: transition probability as a function of the physical distance between neighboring marker sites.