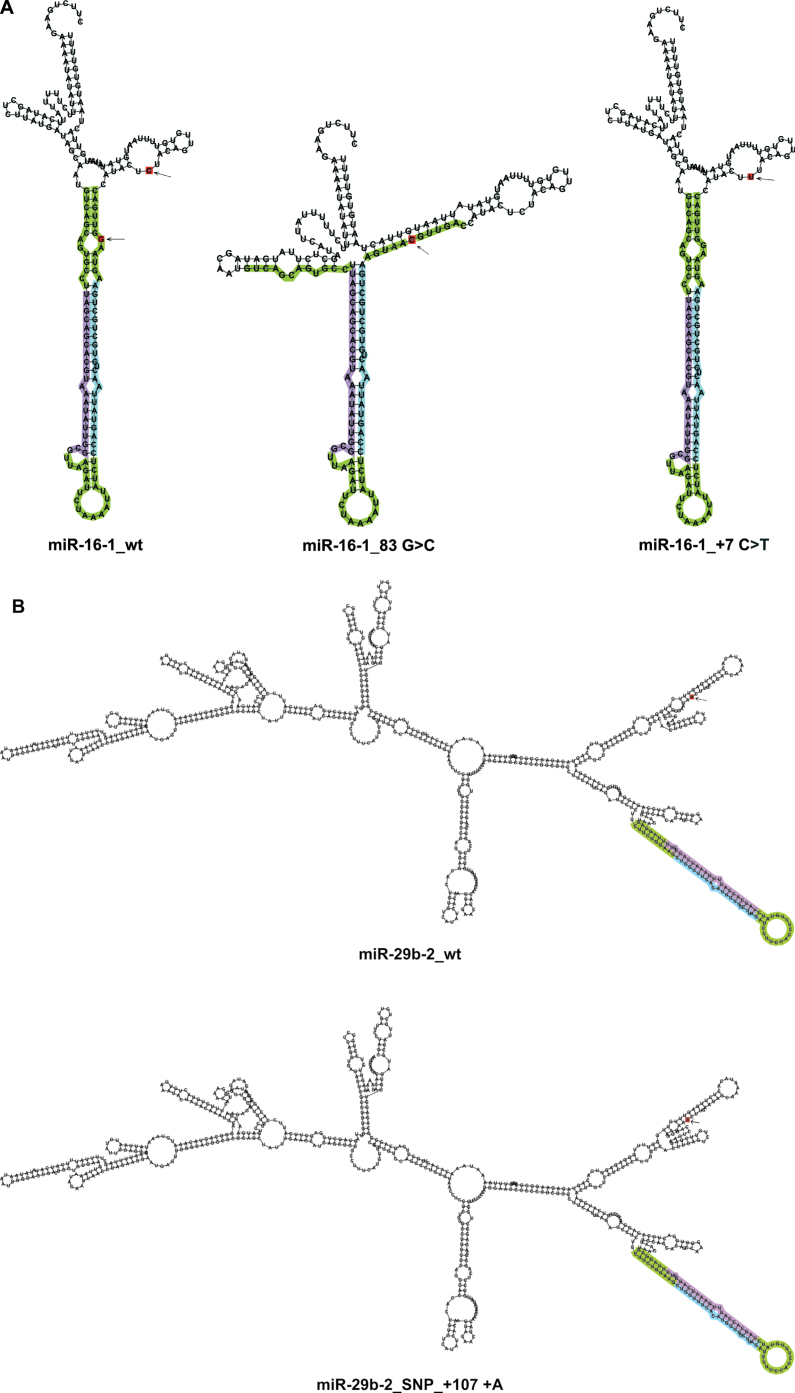
Fig. 4.
Secondary structures of wt miRNAs and miRNAs harboring sequence variations (novel variation/SNP) predicted by the RNAfold tool. (A) The novel variation found in pre-miR-16-1 (83G>C) had a dramatic effect on its secondary structure, whereas the known mutation present in its primary region (+7C>T) (9) had none. (B) pri-miR-29b-2 secondary structure was affected by polymorphic insertion (rs141961287: +107+A).
Depicted are the most stable secondary structures with the lowest free energy as predicted by the RNAfold tool. Variations are designated in red and indicated by arrows. Mature miRNAs and miR-5p are designated in violet; mature miRNAs* and miR-3p are designated in blue. Pre-miRNA regions are designated in green; pri-miRNA regions are colorless.