Abstract
Methylation of cytosine to 5-methylcytosine (5mC) is important for gene expression, gene imprinting, X-chromosome inactivation, and transposon silencing. Active demethylation in animals is believed to proceed by DNA glycosylase removal of deaminated or oxidized 5mC. In plants, 5mC is removed from the genome directly by the DEMETER (DME) family of DNA glycosylases. Arabidopsis thaliana DME excises 5mC to activate expression of maternally imprinted genes. Although the related Repressor of Silencing 1 (ROS1) enzyme has been characterized, the molecular basis for 5mC recognition by DME has not been investigated. Here, we present a structure–function analysis of DME and the related DME-like 3 (DML3) glycosylases for 5mC and its oxidized derivatives. Relative to 5mC, DME and DML3 exhibited robust activity toward 5-hydroxymethylcytosine, limited activity for 5-carboxylcytosine, and no activity for 5-formylcytosine. We used homology modeling and mutational analysis of base excision and DNA binding to identify residues important for recognition of 5mC within the context of DNA and inside the enzyme active site. Our results indicate that the 5mC binding pocket is composed of residues from discrete domains and is responsible for discrimination against 5mC derivatives, and suggest that DME, ROS1, and DML3 utilize subtly different mechanisms to probe the DNA duplex for cytosine modifications.
5-Methylcytosine (5mC) is an important epigenetic modification that serves as a marker for gene expression, X-chromosome inactivation, and transposon silencing, among other developmental processes.1−3 Abnormalities in the regulation of DNA methylation may lead to neuronal disorders and cancer development.4,5 5mC is generated by a class of DNA methyltransferases that use S-adenosylmethionine to methylate cytosine at position C5 by a relatively well characterized mechanism.6 In contrast, the mechanism of 5mC → C demethylation is less well understood. DNA demethylation may occur passively after synthesis of unmethylated daughter strands during replication or actively by enzymes that remove the methylated base. Active demethylation has recently been linked to the base excision repair (BER) pathway, which is normally associated with removal of detrimental nucleobase modifications from the genome.7,8 BER is initiated by DNA glycosylases, which recognize and remove a specifically modified nucleobase by cleaving the N-glycosidic bond reviewed in refs (9−11). In mammals, 5mC may be removed in two ways. In one pathway, activation-induced cytidine deaminase (AID) converts 5mC to thymine, which is excised from the resulting T/G mispair by thymine DNA glycosylase (TDG) and MBD4.12−14 Alternatively, 5mC is oxidized by ten eleven translocation (TET) proteins to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC).15−18 Of these, 5fC and 5caC are substrates for TDG.19,20
In contrast to mammals, plants have evolved specific 5mC DNA glycosylases that remove DNA methylation.21−23Arabidopsis thaliana DEMETER (DME) functions during plant gametogenesis before fertilization and is responsible for imprinting specific genes in the endosperm, which is necessary for seed viability.21,24 DME demethylation in the central and vegetative cells is also thought to produce small interfering RNAs (siRNA) that guide methylation at transposons in the egg and sperm cells, respectively.25,26Arabidopsis also contain three DME paralogs—Repressor of Silencing 1 (ROS1), DME-like 2 (DML2), and DML3—which function in adult cells as genome wide demethylases that remove 5mC marks at sites 3′ and 5′ to genes.27−33 Recent reports of DME activities indicate that the control of DNA methylation can be utilized agriculturally to benefit crop production and medically to develop therapies against celiac disease by suppressing DME expression to produce wheat varieties lacking gliadins and glutenins that cause immunogenic epitopes.34−39
The DME/ROS1/DML enzymes contain a conserved DNA glycosylase domain belonging to the helix-hairpin-helix [4Fe-4S] iron–sulfur cluster superfamily.21 DME and ROS1 utilize a bifunctional glycosylase/lyase mechanism to cleave the glycosidic bond and the phosphodiester backbone through β,δ-elimination to create a one-nucleotide gap in the DNA.22,40 DME/ROS1/DML enzymes are much larger than other glycosylases, ranging from 1100 to over 1700 residues, and contain two additional domains (A and B) with no known homology to other proteins (Figure 1A). Like the glycosylase domain, domains A and B are conserved among the DME paralogs and are essential for DME function.41 Homology modeling of ROS1 predicted a bipartite discontinuous glycosylase domain separated in sequence by an unstructured interdomain region (IDR).42 All DME paralogs contain an N-terminal lysine-rich region that in ROS1 is necessary for binding long stretches of DNA in a methylation-independent manner.21,43
Figure 1.
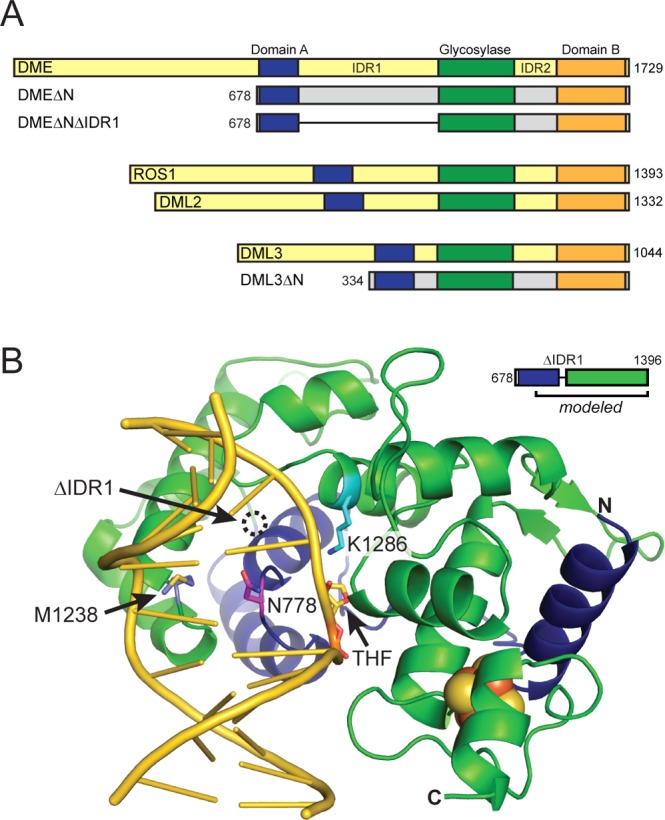
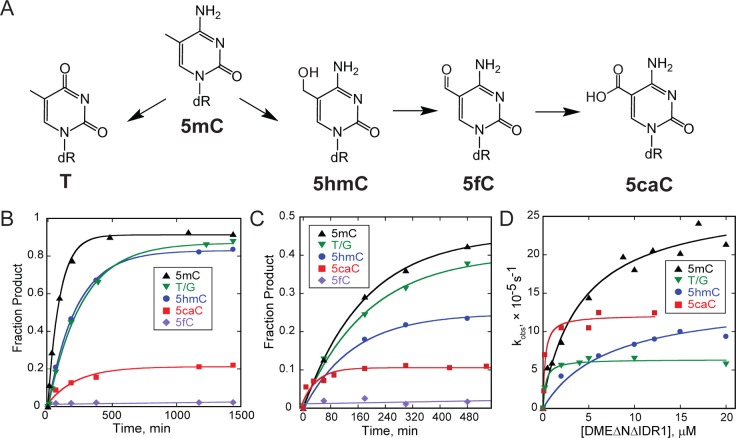
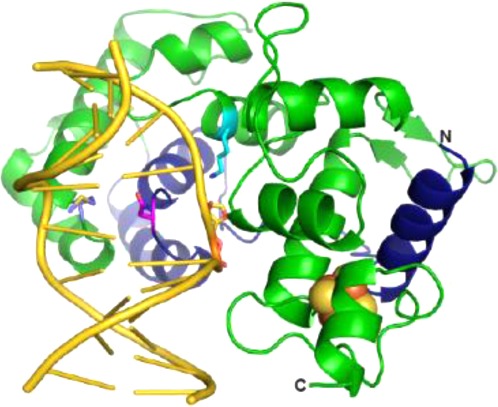
Biochemical characterization of the DME glycosylase domain. (A) Schematic representation of the four Arabidopsis thaliana DME proteins (yellow) and DME and DML3 deletion constructs (gray) used in this study. Conserved domains are indicated by blue (domain A), green (glycosylase), and orange (domain B) boxes. Deletion constructs (ΔN) lack the N-terminal region preceding domain A, and the interdomain region (IDR) 1 has been replaced with a synthetic dodecapeptide linker in DMEΔNΔIDR1. Numbers refer to amino acid positions at the termini. (B) Homology model of DME constructed from the C-terminal half of domain A (residues 737–796) fused to the glycosylase domain (residues 1217–1396). The DNA (gold) is taken from the structure of Nth bound to THF-DNA (PDB ID 1P59), which was used as a template for homology modeling. The location of the Domain A–Glycosylase junction is indicated by the dotted circle. Functionally important residues are represented as sticks (catalytic Lys1286, cyan; plug Asn778, magenta; wedge Met1238, olive).
In contrast to ROS1, which has been the subject of several biochemical studies,42−45 little is known about the molecular mechanisms of DME and DML enzymes. Here, we investigated 5mC excision and substrate specificity of DME and DML3 by mutational analysis. Residues that constitute DME’s active site and DNA damage binding face were predicted from Endonuclease III (EndoIII/Nth) homology modeling, and the importance of these residues to DNA binding and 5mC excision tested. In addition, we previously showed that DME is able to excise 5hmC,46 and we now extend this activity to DML3 and report a comprehensive kinetic analysis of oxidized 5mC excision by DME and DML3. These results provide new insight into the molecular underpinnings for 5mC specificity and excision by the DME family of enzymes.
Materials and Methods
Protein Expression and Purification
Arabidopsis thaliana DMEΔN (residues 678–1729) and DMEΔNΔIDR1, in which residues 798–1188 were replaced with the dodecapeptide AGSSGNGSSGNG, were overexpressed as N-terminal His6-MBP-fusion proteins as described previously.41 The region encoding A. thaliana DML3 amino acids 334–1044 (DML3ΔN) was PCR-amplified from full-length DML3 cDNA template31 using primers 5′-AAT TGG ATC CGT AAC AAC GAT GAT CAA AGC) and 5′-AAT TGA ATT CCT ATA TAT CAT CAT CAC TCA TAA AC. The PCR amplicon was digested with BamHI and EcoRI and cloned into the pET27 (Novagen) derived expression vector pBG102 (Vanderbilt Center for Structural Biology), which produces a cleavable N-terminal His6-SUMO-fusion protein. All constructs were transformed into E. coli Rosetta2 (DE3) cells (Novagen) and propagated in LB media to an OD of 0.5. Proteins were overexpressed for 18 h at 16 °C upon addition of 0.1 mM IPTG. Cells were harvested by centrifugation at 6000 rpm and lysed in 50 mM Tris-HCl pH 8.5, 500 mM NaCl, and 10% glycerol with an Avestin Emulsifier C3 homogenizer operating at ∼20 000 psi. The lysates were cleared by centrifugation at 22 000 rpm for 20 min at 4 °C. Fusion proteins were purified using Ni-NTA (Qiagen) affinity chromatography, followed by cleavage of the affinity tag by PreScission Protease overnight at 4 °C. The proteins were further purified by heparin-sepharose (GE Healthcare) in 50 mM Tris-HCl pH 8.5, 10% glycerol, 0.1 mM DTT, 0.1 mM EDTA, and a NaCl gradient (0.1–1 M), followed by gel filtration on a 16/60 Superdex 200 column (GE Healthcare) in 20 mM Tris-HCl pH 8.5, 200 mM NaCl, 10% glycerol, 0.1 mM DTT, and 0.1 mM EDTA. Protein was concentrated to 100–150 μM and stored at −80 °C in 20 mM Tris-HCl pH 8.5, 150 mM NaCl, 40% glycerol, 0.1 mM DTT, and 0.1 mM EDTA. Mutant proteins were prepared by site-directed mutagenesis using a Quik-Change Kit (Stratagene) and purified in the same manner as the wild-type proteins.
Glycosylase Activity Assay
Proteins were dialyzed in reaction buffer (20 mM Tris-HCl pH 8.5, 150 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA) prior to use. Oligonucleotides of the sequence d(TGACTACTACATGXTTGCCTACCAT), in which X is 5mC, 5hmC, 5fC, 5caC, or T, were synthesized with 6-carboxyfluoroscein at the 5′ end by Integrated DNA Technologies (5mC and T) and Midland Certified Reagents (5hmC, 5fC, and 5caC) and annealed to the complementary strand containing G opposite base X. Enzyme (at least 10 μM for single-turnover conditions) was incubated with 100 nM duplex DNA at 25 °C in reaction buffer. For data shown in Figure 1B, 2 μM protein (nonsaturating) was used. Aliquots of 8 μL were removed from each reaction and terminated with stop buffer (10 mM EDTA in formaldehyde with xylene cyanol and bromophenol blue). Substrate and product DNA were separated by gel electrophoresis with a 20% acrylamide gel in TBE buffer run at 40 W for 1 h. Gels were imaged using a Typhoon Trio variable mode imager and quantified using ImageQuant software (GE Healthcare). Single-turnover rate constants (kst) were determined by exponential fit of the fraction product versus time. To establish the apparent KM (K1/2) for each substrate, rate constants (kobs) were determined at varying enzyme concentrations, and plots of kobs vs [E] were fit to the equation, kobs = kst[E]/(K1/2 + [E]). Under nonsaturating conditions, kobs is dependent on enzyme concentration and is thus reported in units of M–1 s–1.
For inhibition experiments, the glycosylase assay was carried out with 10 μM (5mC excision) and 2 μM (T/G excision) DMEΔNΔIDR1 in the presence of varying concentrations of unlabeled competitor DNA containing tetrahydrofuran (THF), a 1-nucleotide gap, or thymine (T/G mismatch) in the central position. Gap-DNA was assembled by annealing the two 12-mer oligonucleotide sequences flanking the central position to the 25-mer opposite strand. To determine the inhibitory effects of free 5mC or Thy base on 5mC excision, 10 μM DMEΔNΔIDR1 was incubated with varying concentrations of free base (0.01–31 mM) for 30 min prior to the addition of 100 nM fluorescein-labeled 5mC-DNA substrate. The Ki was determined by plotting kobs as a function of competitor DNA concentration and fitting the data to the equation, kobs = kst[E]/(Ki + [E]).
DNA Binding
Oligonucleotides containing the same sequence as above and a centrally located 5mC were annealed to complementary strand containing 6-carboxyfluorescein at the 3′-end. Varying concentrations of enzyme (0–100 μM) were incubated with 50 nM duplex DNA in reaction buffer for 10 min at 25 °C. Fluorescence anisotropy was measured using a Biotek Synergy H1 hybrid multimode microplate reader at excitation and emission wavelengths of 485 and 538 nm, respectively. Dissociation constants (Kd) were calculated by fitting the data to a two-state binding model. Reported values are averages from three independent experiments. Because DME reacts slowly, there is no significant product generated within 10 min. Anisotropy values measured 0, 10, and 30 min after addition of wild-type enzyme did not change.
DME Homology Model
A homology model of DME bound to abasic-DNA was constructed using Swiss-Model47 and the structure of Bacillus stearothermophilus EndoIII/Nth (PDB ID 1P59(48)) as a template, similar to that previously described for ROS1.42 The final model incorporated DME residues 737–796 of domain A and 1217–1396 of the glycosylase domain. Protein–DNA contacts were predicted using the coordinates of the tetrahydrofuran (THF)-containing DNA from the Nth structure superimposed on the DME model.
Results
Model for DNA Binding by DME
Arabidopsis DME, ROS1, DML2, and DML3 each contain three highly conserved domains important for function (Figure 1A). The N-terminal and interdomain regions (IDRs) have very low homology and little predicted secondary structure. We previously showed that the three conserved domains are necessary and sufficient for DME 5mC excision activity, as constructs lacking the N-terminal 677 residues (DMEΔN) and with the IDR1 replaced with a short dodecapeptide linker (DMEΔNΔIDR1) retain activity.41 In order to quantify the effect of IDR1 removal on enzymatic activity, we compared the kinetics of 5mC excision of DMEΔNand DMEΔNΔIDR1 proteins. Under nonsaturating conditions, the observed rate constants (kobs) of DMEΔNand DMEΔNΔIDR1 are 134 ± 9 M–1 s–1 and 43 ± 1 M–1 s–1, respectively (Figure S1). Thus, removal of the 393-residue region between domain A and glycosylase domain has only a 3-fold effect on the rate of 5mC excision, which, under the conditions of this assay, is not significant. However, deletion of IDR1 significantly improves protein solubility, purity, and stability, mainly as a result of proteolytic sensitivity of IDR1, making DMEΔNΔIDR1 more amenable to biochemical analysis than DME or DMEΔN. Therefore, the DMEΔNΔIDR1 construct was used for the remainder of this study.
In order to gain insight into the DME structural features important for 5mC excision, we constructed a homology model of the glycosylase domain using the structure of Bacillus stearothermophilus Nth, a closely related [4Fe-4S]-containing glycosylase specific for oxidized purines. As reported previously for ROS1,42 the C-terminal region (residues 737–796) of domain A is predicted to complete the helix-hairpin-helix glycosylase fold based on its significant sequence conservation with the N-terminal 60 residues of Nth (Figure 1C). The conserved regions place DME Phe796 (domain A) in close proximity to DME Asp1217 (glycosylase domain) such that IDR1 is excluded entirely and most likely an independent domain. This aspect of the model is validated by the retention of activity in the DMEΔNΔIDR1 mutant and the inclusion of functionally important residues within the region contributed by domain A.41 In addition, the model places the catalytic Asp1304 and Lys1286 in close proximity to the flipped abasic site. Substitution of either of these residues (Asp1304Asn and Lys1286Gln) abrogates 5mC excision activity (Table 1).22
Table 1. DNA Binding and 5mC Excision Activities for DMEΔNΔIDR1 Point Mutantsa.
| Kd (μM) | relative affinity | kst (×10–5 s–1) | relative to WT | |
|---|---|---|---|---|
| WT | 8.3 ± 1.2 | 1.0 | 18.1 ± 1.2 | 1.0 |
| Q777A | 8.1 ± 1.1 | 1.0 | 7.6 ± 0.4 | 0.4 |
| N778A | 18.9 ± 3.4 | 0.4 | 2.7 ± 0.6 | 0.1 |
| M1238A | 8.5 ± 1.0 | 1.0 | 0.02 ± 0.01 | 0.001 |
| K1286Q | 8.2 ± 1.8 | 1.0 | <0.01b | 0.0005 |
| D1304N | 0.6 ± 0.1 | 14 | <0.01b | 0.0005 |
Dissociation constants (Kd) and single-turnover rate constants for 5mC (kst) excision were measured at 25 °C, pH 8.5, and 170 mM ionic strength using a 25mer DNA duplex containing 5mC and end-labeled with fluorescein. Enzyme concentration was saturating (10 μM) in activity assays. Values are reported as averages ± SD from at least three experiments. Relative binding affinities calculated as (Kd WT)/(Kd Mutant); Relative to WT rates calculated as (kst Mutant)/(kst WT).
Activity is below limit of detection.
Damage recognition and catalysis by DNA glycosylases is facilitated by extrusion of the target nucleobase inside an active site pocket. This extrahelical conformation is stabilized by intercalating side chains that plug the gap left by the flipped nucleobase and create a wedge in the opposite strand to create a sharp kink in the DNA duplex.9−11,49−52 Substitution of these intercalating residues typically has a dramatic effect on glycosylase activity. Our homology model predicts that Asn778 and Met1238 would serve as the plug and wedge residues in DME (Figure 1C). Substitution of Met1238 with alanine in DMEΔNΔIDR1 abrogated 5mC excision activity and had no effect on DNA binding (Figures 1D and S2, Table 1), consistent with results from the corresponding residue (Met905) in ROS1 (Figure S2).42,45 Similarly, an Asn778Ala substitution had a 10-fold reduction in activity relative to WT (Figure 1D, Table 1). Interestingly, the corresponding substitution in ROS1 (Asn608Ala) had no effect.42 Rather, the neighboring ROS1 residue (Gln607) was found to be necessary for 5mC removal and duplex interrogation.42,45 In contrast, substitution of this neighboring residue in DME (Gln777) with alanine had only a 2-fold effect on 5mC excision and no effect on DNA binding relative to wild-type (Figures 1D and S2, Table 1). We also tested these two putative plug residues in DML3ΔN by making alanine substitutions corresponding to DME Gln777 and Asn778. In this case, both DML3 Gln426Ala and Asn427Ala abrogated 5mC excision activity (Figure S2), suggesting that DME and DML3 utilize subtly different modes of damage recognition. Thus, we have identified DME and DML3 residues from discrete domains as likely candidates for duplex interrogation and/or stabilization of extrahelical 5mC.
Specificity of DME and DML3 for Oxidized 5mC
In mammals, 5mC is oxidized to 5hmC, 5fC, and 5caC by TET proteins15−18 (Figure 2A). We recently found that DME and ROS1 have activity toward 5hmC.46 Although we were unable to detect these derivatives in Arabidopsis, we compared their rates of excision by DMEΔNΔIDR1 and DML3ΔN against 5mC and thymine from T/G-mismatches as a means to understand the molecular basis for the unique 5mC specificity of the DME family of enzymes. Under single-turnover conditions, DME exhibited the same activity for T/G and 5hmC, with first-order rate constants (kst) only 2–3-fold less than for 5mC excision (Figure 2B, Table 2). Interestingly, the rate of 5caC excision was also 2-fold less than 5mC (Table 2), although the enzyme was only able to process 20% of the substrate (Figure 2B), suggesting either a mixture of 5caC species (e.g., amino and imino forms)53 was present in the DNA or a product of 5caC excision inhibited the reaction. Under the same conditions, DME was inactive toward 5fC (Figure 2B). DML3 excised 5mC 2-fold more slowly than DME and showed the same general trend toward excision of oxidized 5mC derivatives (Figure 2C and Table 2).
Figure 2.
Excision of oxidized 5mC nucleobases by DME and DML3. (A) 5mC can be deaminated to form thymine or successively oxidized to 5hmC, 5fC, and 5caC. (B,C) Glycosylase activities of DMEΔNΔIDR1 (B) and DML3ΔN(C) used to determine single-turnover rates (kst) shown in Table 2. (D) DMEΔNΔIDR1 concentration dependence on rates of 5mC, 5hmC, 5caC, and T/G excision used to determine K1/2 values shown in Table 3.
Table 2. Rates of Excision of Oxidized 5mC Derivatives by DME and DML3a.
| 5mC | 5hmC | 5fC | 5caCb | T/G | |
|---|---|---|---|---|---|
| DMEΔNΔIDR1 | 18.1 ± 1.2 | 8.3 ± 1.1 | 0.02 ± 0.01 | 8.4 ± 1.6 | 6.5 ± 0.6 |
| DML3ΔN | 9.1 ± 0.3 | 11.4 ± 5.3 | 0.04 ± 0.02 | 35 ± 10 | 9.2 ± 0.6 |
Single-turnover rate constants (kst, ×10–5 s–1) for excision of oxidized 5mC derivatives from a 25mer oligonucleotide at 25 °C, pH 8.5, 170 mM ionic strength, and 10 μM enzyme. Values are reported as averages ± SD from at least three experiments.
Enzyme removes only 20% 5caC.
In order to establish the preferred substrate of DME, we determined the apparent KM (K1/2) using the DMEΔNΔIDR1 construct. Interestingly, the preference of DME for each substrate varied greatly (Figure 2D). The K1/2 for 5hmC was comparable to 5mC, whereas K1/2 values for 5caC and T/G were at least 25-fold smaller, indicating a preference for binding these substrates (Table 3). However, the kst for 5hmC, 5caC, and T/G were all lower than 5mC, suggesting inhibition by an intermediate or product of these reactions (Figure 2D). Because DNA glycosylases are often product inhibited and abasic sites opposite 5mC inhibit DME activity,22 we tested the inhibitory effects of DNA containing a THF abasic site mimetic or a 1-nucleotide gap, which resemble the reaction intermediate and product, respectively, on 5mC and T/G excision activity by DME (Figure S4). The abasic and gapped DNA products inhibited 5mC excision with Ki values of 11.3 ± 4.3 μM and 12.6 ± 3.3 μM, respectively, which were roughly equivalent to the enzyme concentration used (10 μM) and to the strength of competition by 5mC-DNA (Ki = 4.3 ± 2.1 μM) and T/G-DNA (Ki = 7.7 ± 3.3 μM) substrates (Figure S4A,B). Similarly, the DNAs inhibited T/G excision with Ki values of 2.4 ± 0.6 μM (THF-DNA) and 6.8 ± 0.8 μM (Gap-DNA) (Figure S4C), similar to the enzyme concentration used (2 μM). Therefore, the DNA products inhibited DME excision of 5mC and T/G to the same extent. Consistent with this, Kd values measured by fluorescence anisotropy for binding of the catalytically dead D1304N mutant to DNA containing 5mC and T/G were 0.6 ± 0.1 μM and 0.2 ± 0.1 μM, respectively (Figure S3). In contrast to DNA intermediates and products, we were unable to inhibit 5mC excision by free nucleobases, even at high concentrations (Figure S4D).
Table 3. Substrate Specificity of DMEa.
| substrate | K1/2, μM |
|---|---|
| 5mC | 5.1 ± 1.7 |
| 5hmC | 7.4 ± 0.8 |
| 5fC | NDb |
| 5caC | 0.2 ± 0.9 |
| T/G | 0.12 ± 0.04 |
Values are reported as averages ± SD from at least three experiments.
ND, not determined.
Active Site Residues Confer Specificity for Substrate DNA
Substrate specificity of the HhH superfamily of DNA glycosylases is largely determined by the shape and chemical complementarity between the nucleobase within the active site pocket.11 The active site of DME predicted by the homology model is formed by residues contributed by both A and glycosylase domains (Figure 1A), similar to that previously shown for ROS1.42 Superimposing an extrahelical 5mC onto our homology model and an examination of other glycosylase–DNA structures, including MBD4 in complex with T/G-DNA (PDB ID 4EVV), led to the identification of six residues lining the DME nucleobase binding pocket: Phe759, Thr776, and Asp781 from Domain A; and His1360, Tyr1361, and Ile1364 from the glycosylase domain (Figure 3A). Phe759 is predicted to reside on an unstructured loop and to stack against the N4 nitrogen of 5mC. Thr776 and Asp781 sit adjacent to the abasic site DNA in the DME homology model and correspond to two MBD4 residues that were observed to make contacts to the flipped thymine substrate in the crystal structures of MBD4 bound to DNA.54−56 His1360, Tyr1361, and Ile1364 reside near the predicted location of 5mC in the active site.
Figure 3.
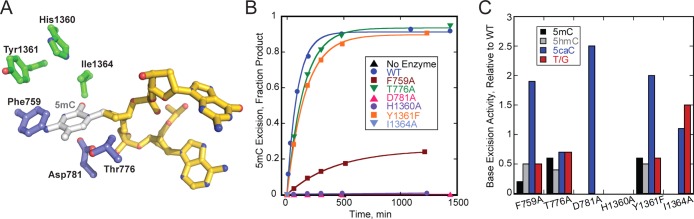
DME active site mutants affect base excision. (A) Homology model of DME’s active site. Protein residues are colored by domain as in Figure 1. THF-containing DNA (gold) is modeled from the structure of Nth bound to THF-DNA (PDB ID 1P59), and a modeled 5mC is shown in silver. (B) Single-turnover excision rates of 5mC by DME active site mutants compared with wild-type (WT) DMEΔNΔIDR1. (C) Single-turnover rates of 5mC, 5hmC, 5caC, or T/G excision by each DME active site mutant relative to wild-type.
We examined the contribution of each predicted active site residue to DME’s substrate specificity by mutational analysis of 5mC, 5hmC, 5caC, and T/G excision activity using the DMEΔNΔIDR1 construct (Figure 3B,C). All mutants purified well and retained the brilliant yellow color of wild-type DME, indicating proper folding of the proteins. Thr776Ala had only a modest reduction in 5mC excision activity relative to wild-type. In contrast, leucine substitution of the corresponding residue in ROS1 (Thr606) abolished ROS1 activity.42 DME activity also differed from ROS1 with respect to Tyr1361 (Tyr1028 in ROS1). DME Tyr1361Phe reduced 5mC and T/G excision activity nearly 2-fold, whereas ROS1 Tyr1028Ser reduced 5mC activity 2-fold but increased preference for T/G excision.42 In contrast to Thr776Ala and Tyr1361Phe, which had only a modest effect on 5mC excision by DME, Phe759Ala significantly reduced 5mC excision, and Asp781Ala, His1360Ala, and Ile1364Ala abrogated 5mC excision (Figure 3B,C). A similar reduction in 5mC activity was observed for ROS1 Asp611Val, which corresponds to DME Asp781.42 Similarly, DME Phe759Ala showed a higher activity for T/G excision than 5mC excision, consistent with the corresponding residue in ROS1 (Phe589Ala).42 His1360Ala was the only mutation in DME that abrogated excision of all substrates (Figures 3C and S5). Although Ile1364Ala had no activity for 5mC or 5hmC, it retained activity for 5caC and T/G at rates similar to wild-type (Figure 3C and Table S1). Surprisingly, three mutations (Phe759Ala, Asp781Ala, and Tyr1361Phe) showed a modest increase in 5caC excision rates relative to wild-type, but remained unable to excise more than 20% of the 5caC-DNA substrate (Figures 3C and S5). Interestingly, the Asp781Ala substitution showed the highest increase in 5caC activity despite complete loss of activity toward the other nucleobases tested. Taken together, these results validate the homology model, identify His1360, Asp781, and Ile1364 as important for DME activity, and suggest that the various 5mC derivatives may be recognized through architectural changes to the active site pocket.
Discussion
This work represents the first quantitative structure–function study of DME and DML3, providing a molecular rationale for their similarities and differences to ROS1, which has been extensively characterized.42−45 We previously identified the catalytic core of DME to be composed of domains A and B in addition to the glycosylase domain and found that IDR1 is dispensable for 5mC excision.41 Here, using the structure of the Nth-DNA complex as a guide, homology modeling of DME revealed that domain A contributes three α-helices to the glycosylase domain as previously identified in ROS1.42 The model also predicts that the IDR1 insertion connects to the glycosylase domain on the side opposite the DNA binding site, consistent with the fact that deletion of IDR1 had only a modest effect on DME glycosylase activity in vitro. Moreover, the rate of 5mC excision by DME was comparable to DML3 (Table 1) and ROS1,44 which contain variable IDR1s in both length and sequence, further implying that the IDRs are not critical for 5mC excision activity. This large insertion to a glycosylase fold is unique to the DME family, leading us to speculate that these large insertions function to regulate in vivo activity, possibly by mediating protein interactions. For example, specific IDR1–protein interactions may facilitate DME’s activation of imprinted genes at specific loci and ROS1 and DML3’s genome-wide demethylase activity.27−30
Our homology model and mutagenesis results indicate that Asn778 and Met1238 are plug and wedge residues, respectively, important for 5mC detection. These two residues were identified to be critical for DME activity from previous random mutagenesis experiments, which were performed in vivo in the context of full-length DME.41 Interestingly, despite the similarity in their predicted structures, DME, ROS1, and DML3 seem to utilize different duplex interrogating residues. All DME family members contain an Asn-Gln moiety predicted to reside on the loop containing the plug residue in other glycosylases (Figure S2). Whereas alanine substitution of the Gln had a 10-fold greater effect than that of the Asn residue on ROS1 activity, we found the opposite to be true for DME. Moreover, alanine substitution of either Asn or Gln abolished DML3 activity. These data suggest that the DNA intercalation loop in each DME paralog engages the DNA differently. Because the residues from this loop have been shown to be important for interrogation of DNA during discrimination of modified versus unmodified nucleobases prior to base flipping,57 a different mode of intercalation would likely alter the mechanism by which each enzyme locates its 5mC target. Moreover, this interrogating loop in HhH glycosylases plays a role in recognition of the base opposite the lesion. For example, Nth contacts the opposite base through hydrogen bonds from the backbone carbonyl of Gln42,48 which aligns with DME Asn778. There are no other candidate interactions with the opposing G in our homology model, and thus we do not expect there to be a strong opposite base specificity in DME. Consistent with this, ROS1 excises 5mC more efficiently when mispaired with C, T, or A, which is likely due to the reduced thermodynamic stability of the base pair.44
Despite differences in the identity of the plug residue, both DME and ROS1 utilize a conserved Met residue from the glycosylase domain to wedge between the nucleobases opposite 5mC.42 Furthermore, we observed both similarities and differences between the predicted active sites of DME and ROS1. Most notably, the residue corresponding to DME Thr776 is essential to 5mC excision in ROS1,42 but not in DME. Likewise, DME Tyr1361Phe did not cause a change in substrate specificity between 5mC and T/G, although the corresponding mutation in ROS1 did.42 Therefore, despite the similarity among the DME family of enzymes, we find subtle differences in substrate recognition by both DNA intercalation and active site residues that may help explain the differences in sites of demethylation in the Arabidopsis genome.27−33
We found the order of substrate preference for DME and DML3 to be 5mC > 5hmC ∼ T/G > 5caC > 5fC. However, when comparing the rates of excision, it appeared that DME bound both 5caC and T/G tighter than 5mC and even excised 5caC faster than 5mC at low levels of enzyme. DME was not inhibited by free 5mC or thymine base, and thus the low maximal rate of T/G excision compared to 5mC cannot be explained by product thymine remaining in the active site. In contrast, we found that both 5mC and T/G excision were inhibited in the presence of product-DNA. However, less product-DNA was needed to reduce T/G excision, indicating a higher affinity of DME for 5mC-DNA as compared with T/G-DNA. Therefore, we conclude that the apparent high affinity of DME for T/G- and 5caC-DNA can be attributed to product DNA inhibition and to the slow turnover of the enzyme for these substrates, as evidenced by their low kst values. Release of the abasic DNA product is often the rate-limiting step in glycosylase reactions and is promoted by AP-endonuclease, the next enzyme in BER pathway.58−61 Consistent with this interpretation, ROS1 excises 5mC and binds to the resulting abasic site in a distributive manner.44
DNA glycosylases, including those belonging to the helix-hairpin-helix superfamily, are either specific for oxidation or alkylation damage, but not both.11 The ability of the DME family of enzymes to remove both methylated 5mC and oxidized 5hmC makes them unusual. This unique specificity is important in light of the recent progress to understand active demethylation by TDG in mammals.62 While DME excises 5hmC relatively well compared to 5mC, it does not effectively remove 5fC or 5caC. In contrast, TDG shows no activity for 5hmC, and has 40% greater activity for 5fC and 25% residual activity for 5caC relative to its preferred T/G substrate. Thus, DME and TDG have reciprocal specificities with respect to the oxidized derivatives that may reflect the fact that the 5mC oxidation pathway is not needed in plants. Although there is no evidence to suggest that oxidized 5mC nucleobases are biologically relevant substrates in Arabidopsis,46 the ability of DME to remove these bases provides insight into 5mC discrimination inside the binding pocket. Residues contributed by both domain A (Phe759 and Asp781) and the glycosylase domain (His1360 and Ile1364) are necessary for full 5mC excision activity. These data are consistent with our previous random mutagenesis screen, in which mutations of Asp781 and His1360 abolished DME activity.41 Similarly, the lack of an effect of Thr776Ala or Tyr1361Phe on 5mC excision correlates with the absence of Thr776 or Tyr1361 substitutions from the random screen. Interestingly, Ile1364 abolished 5mC and 5hmC excision, but did not affect activity toward T/G, implicating this residue as an important recognition element of cytosine derivatives. Phe579, located in close proximity to the 5mC methyl substituent, was the only residue to show a difference, albeit modest, between 5mC and 5hmC activity. In contrast, the relatively large distance of His1360 from the flipped base suggests that substitution of this residue affected the folding of the binding pocket. Although these data start to paint a picture of the 5mC active site, it is conceivable that domain B, lying outside of the nucleobase binding pocket, contributes to 5mC recognition, similar to the 8-oxoguanine recognition domain of MutY.9 Indeed, domain B contains residues essential to DME activity.41 Further structural analysis will be necessary to elucidate the roles of domains A and B in DME.
Acknowledgments
The authors thank the members of the Eichman lab for helpful suggestions and advice.
Glossary
Abbreviations
- DME
DEMETER
- 5mC
5-methylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxylcytosine
- THF
tetrahydrofuran
- EndoIII/Nth
endonuclease III
Supporting Information Available
Table S1, Figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by the National Institutes of Health (ES019625 to B.F.E., GM69415 to R.L.F.) and the Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea (PJ009092 to J.H.H.). S.C.B. was supported by a National Science Foundation Graduate Research Fellowship (0909667) and the Vanderbilt Molecular Biophysics Training Grant from the National Institutes of Health (T32 GM08320). Additional support for facilities was provided by National Institutes of Health funding to the Vanderbilt Center in Molecular Toxicology (P30 ES000267) and the Vanderbilt-Ingram Cancer Center (P30 CA068485).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Chan S. W.; Henderson I. R.; Jacobsen S. E. (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351–360. [DOI] [PubMed] [Google Scholar]
- Huh J. H.; Bauer M. J.; Hsieh T. F.; Fischer R. L. (2008) Cellular programming of plant gene imprinting. Cell 132, 735–744. [DOI] [PubMed] [Google Scholar]
- Law J. A.; Jacobsen S. E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. (2010) Brain function and chromatin plasticity. Nature 465, 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P.; Ohlsson R.; Henikoff S. (2006) The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7, 21–33. [DOI] [PubMed] [Google Scholar]
- Goll M. G.; Bestor T. H. (2005) Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514. [DOI] [PubMed] [Google Scholar]
- Piccolo F. M.; Fisher A. G. (2013) Getting rid of DNA methylation. Trends Cell Biol. 24, 136–143. [DOI] [PubMed] [Google Scholar]
- Lindahl T. (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res. 462, 129–135. [DOI] [PubMed] [Google Scholar]
- Fromme J. C.; Banerjee A.; Huang S. J.; Verdine G. L. (2004) Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427, 652–656. [DOI] [PubMed] [Google Scholar]
- Huffman J. L.; Sundheim O.; Tainer J. A. (2005) DNA base damage recognition and removal: new twists and grooves. Mutat. Res. 577, 55–76. [DOI] [PubMed] [Google Scholar]
- Brooks S. C.; Adhikary S.; Rubinson E. H.; Eichman B. F. (2013) Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta 1834, 247–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S.; Xu J.; Sannai M.; Moore R.; Caretti E.; Cigliano A.; Le Coz M.; Devarajan K.; Wessels A.; Soprano D.; Abramowitz L. K.; Bartolomei M. S.; Rambow F.; Bassi M. R.; Bruno T.; Fanciulli M.; Renner C.; Klein-Szanto A. J.; Matsumoto Y.; Kobi D.; Davidson I.; Alberti C.; Larue L.; Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar D.; Kunz C.; Selfridge J.; Lettieri T.; Saito Y.; MacDougall E.; Wirz A.; Schuermann D.; Jacobs A. L.; Siegrist F.; Steinacher R.; Jiricny J.; Bird A.; Schar P. (2011) Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423. [DOI] [PubMed] [Google Scholar]
- Rai K.; Huggins I. J.; James S. R.; Karpf A. R.; Jones D. A.; Cairns B. R. (2008) DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S.; Shen L.; Dai Q.; Wu S. C.; Collins L. B.; Swenberg J. A.; He C.; Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M.; Koh K. P.; Shen Y.; Pastor W. A.; Bandukwala H.; Brudno Y.; Agarwal S.; Iyer L. M.; Liu D. R.; Aravind L.; Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron N.; Peters A. H. (2011) Epigenetics: Tet proteins in the limelight. Nature 473, 293–294. [DOI] [PubMed] [Google Scholar]
- Wu H.; Zhang Y. (2011) Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 25, 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A.; Drohat A. C. (2011) Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286, 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Lu X.; Lu J.; Liang H.; Dai Q.; Xu G. L.; Luo C.; Jiang H.; He C. (2012) Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 8, 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.; Gehring M.; Johnson L.; Hannon M.; Harada J. J.; Goldberg R. B.; Jacobsen S. E.; Fischer R. L. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110, 33–42. [DOI] [PubMed] [Google Scholar]
- Gehring M.; Huh J. H.; Hsieh T. F.; Penterman J.; Choi Y.; Harada J. J.; Goldberg R. B.; Fischer R. L. (2006) DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Ruiz T.; Ortega-Galisteo A. P.; Ponferrada-Marin M. I.; Martinez-Macias M. I.; Ariza R. R.; Roldan-Arjona T. (2006) DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. U. S. A. 103, 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. F.; Shin J.; Uzawa R.; Silva P.; Cohen S.; Bauer M. J.; Hashimoto M.; Kirkbride R. C.; Harada J. J.; Zilberman D.; Fischer R. L. (2011) Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U. S. A. 108, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco J. P.; Borges F.; Donoghue M. T.; Van Ex F.; Jullien P. E.; Lopes T.; Gardner R.; Berger F.; Feijo J. A.; Becker J. D.; Martienssen R. A. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151, 194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra C. A.; Feng X.; Schoft V. K.; Hsieh T. F.; Uzawa R.; Rodrigues J. A.; Zemach A.; Chumak N.; Machlicova A.; Nishimura T.; Rojas D.; Fischer R. L.; Tamaru H.; Zilberman D. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D.; Gehring M.; Tran R. K.; Ballinger T.; Henikoff S. (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39, 61–69. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Yazaki J.; Sundaresan A.; Cokus S.; Chan S. W.; Chen H.; Henderson I. R.; Shinn P.; Pellegrini M.; Jacobsen S. E.; Ecker J. R. (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–1201. [DOI] [PubMed] [Google Scholar]
- Gong Z.; Morales-Ruiz T.; Ariza R. R.; Roldan-Arjona T.; David L.; Zhu J. K. (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814. [DOI] [PubMed] [Google Scholar]
- Zhu J.; Kapoor A.; Sridhar V. V.; Agius F.; Zhu J. K. (2007) The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 17, 54–59. [DOI] [PubMed] [Google Scholar]
- Penterman J.; Zilberman D.; Huh J. H.; Ballinger T.; Henikoff S.; Fischer R. L. (2007) DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U. S. A. 104, 6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J.; Uzawa R.; Fischer R. L. (2007) Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 145, 1549–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo A. P.; Morales-Ruiz T.; Ariza R. R.; Roldan-Arjona T. (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681. [DOI] [PubMed] [Google Scholar]
- Kapazoglou A.; Drosou V.; Argiriou A.; Tsaftaris A. S. (2013) The study of a barley epigenetic regulator, HvDME, in seed development and under drought. BMC Plant Biol. 13, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y.; Kwak K. J.; Jung H. J.; Lee H. J.; Kang H. (2010) MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol. 51, 1079–1083. [DOI] [PubMed] [Google Scholar]
- Osorio C.; Wen N.; Gemini R.; Zemetra R.; von Wettstein D.; Rustgi S. (2012) Targeted modification of wheat grain protein to reduce the content of celiac causing epitopes. Funct. Integr. Genomics 12, 417–438. [DOI] [PubMed] [Google Scholar]
- Mitea C.; Salentijn E. M.; van Veelen P.; Goryunova S. V.; van der Meer I. M.; van den Broeck H. C.; Mujico J. R.; Montserrat V.; Gilissen L. J.; Drijfhout J. W.; Dekking L.; Koning F.; Smulders M. J. (2010) A universal approach to eliminate antigenic properties of alpha-gliadin peptides in celiac disease. PLoS One 5, e15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi P. F.; Wei Y. M.; Ouellet T.; Chen Q.; Tan X.; Zheng Y. L. (2009) The gamma-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genomics 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S.; Wen N.; Pang J.; Langen G.; Brew-Appiah R. A.; Mejias J. H.; Osorio C.; Yang M.; Gemini R.; Moehs C. P.; Zemetra R. S.; Kogel K. H.; Liu B.; Wang X.; von Wettstein D.; Rustgi S. (2012) Structural genes of wheat and barley 5-methylcytosine DNA glycosylases and their potential applications for human health. Proc. Natl. Acad. Sci. U. S. A. 109, 20543–20548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agius F.; Kapoor A.; Zhu J. K. (2006) Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. U. S. A. 103, 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok Y. G.; Uzawa R.; Lee J.; Weiner G. M.; Eichman B. F.; Fischer R. L.; Huh J. H. (2010) Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc. Natl. Acad. Sci. U. S. A. 107, 19225–19230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marin M. I.; Parrilla-Doblas J. T.; Roldan-Arjona T.; Ariza R. R. (2011) A discontinuous DNA glycosylase domain in a family of enzymes that excise 5-methylcytosine. Nucleic Acids Res. 39, 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marin M. I.; Martinez-Macias M. I.; Morales-Ruiz T.; Roldan-Arjona T.; Ariza R. R. (2010) Methylation-independent DNA binding modulates specificity of Repressor of Silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 285, 23032–23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marin M. I.; Roldan-Arjona T.; Ariza R. R. (2009) ROS1 5-methylcytosine DNA glycosylase is a slow-turnover catalyst that initiates DNA demethylation in a distributive fashion. Nucleic Acids Res. 37, 4264–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Doblas J. T.; Ponferrada-Marin M. I.; Roldan-Arjona T.; Ariza R. R. (2013) Early steps of active DNA demethylation initiated by ROS1 glycosylase require three putative helix-invading residues. Nucleic Acids Res. 41, 8654–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Shin H., Eichman B. F., and Huh J. H.. Excision of 5-hydroxymethylcytosine by DEMETER family DNA glycosylases, Biochem. Biophys. Res. Commun. 2014, 10.1016/j.bbrc2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N.; Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Fromme J. C.; Verdine G. L. (2003) Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 22, 3461–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B.; Laerdahl J. K.; Backe P. H.; Bjoras M. (2009) DNA base repair--recognition and initiation of catalysis. FEMS Microbiol. Rev. 33, 1044–1078. [DOI] [PubMed] [Google Scholar]
- David S. S.; O’Shea V. L.; Kundu S. (2007) Base-excision repair of oxidative DNA damage. Nature 447, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. I.; Stivers J. T. (2010) Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 49, 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. M. (2010) Novel molecular insights into the mechanism of GO removal by MutM. Cell Res. 20, 116–118. [DOI] [PubMed] [Google Scholar]
- Maiti A.; Michelson A. Z.; Armwood C. J.; Lee J. K.; Drohat A. C. (2013) Divergent mechanisms for enzymatic excision of 5-formylcytosine and 5-carboxylcytosine from DNA. J. Am. Chem. Soc. 135, 15813–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J.; Arita K.; Kato T.; Kinoshita M.; Kimura H.; Suetake I.; Tajima S.; Ariyoshi M.; Shirakawa M. (2013) Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J. Biol. Chem. 288, 6351–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manvilla B. A.; Maiti A.; Begley M. C.; Toth E. A.; Drohat A. C. (2012) Crystal structure of human methyl-binding domain IV glycosylase bound to abasic DNA. J. Mol. Biol. 420, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morera S.; Grin I.; Vigouroux A.; Couve S.; Henriot V.; Saparbaev M.; Ishchenko A. A. (2012) Biochemical and structural characterization of the glycosylase domain of MBD4 bound to thymine and 5-hydroxymethyuracil-containing DNA. Nucleic Acids Res. 40, 9917–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.; Santos W. L.; Verdine G. L. (2006) Structure of a DNA glycosylase searching for lesions. Science 311, 1153–1157. [DOI] [PubMed] [Google Scholar]
- Baldwin M. R.; O’Brien P. J. (2010) Nonspecific DNA binding and coordination of the first two steps of base excision repair. Biochemistry 49, 7879–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters T. R.; Gallinari P.; Jiricny J.; Swann P. F. (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 274, 67–74. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. E.; Drohat A. C. (2008) Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J. Biol. Chem. 283, 32680–32690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko V. S.; Nevinsky G. A.; Zharkov D. O. (2007) Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair 6, 317–328. [DOI] [PubMed] [Google Scholar]
- Wu H.; Zhang Y. (2014) Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156, 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.