Figure 7.
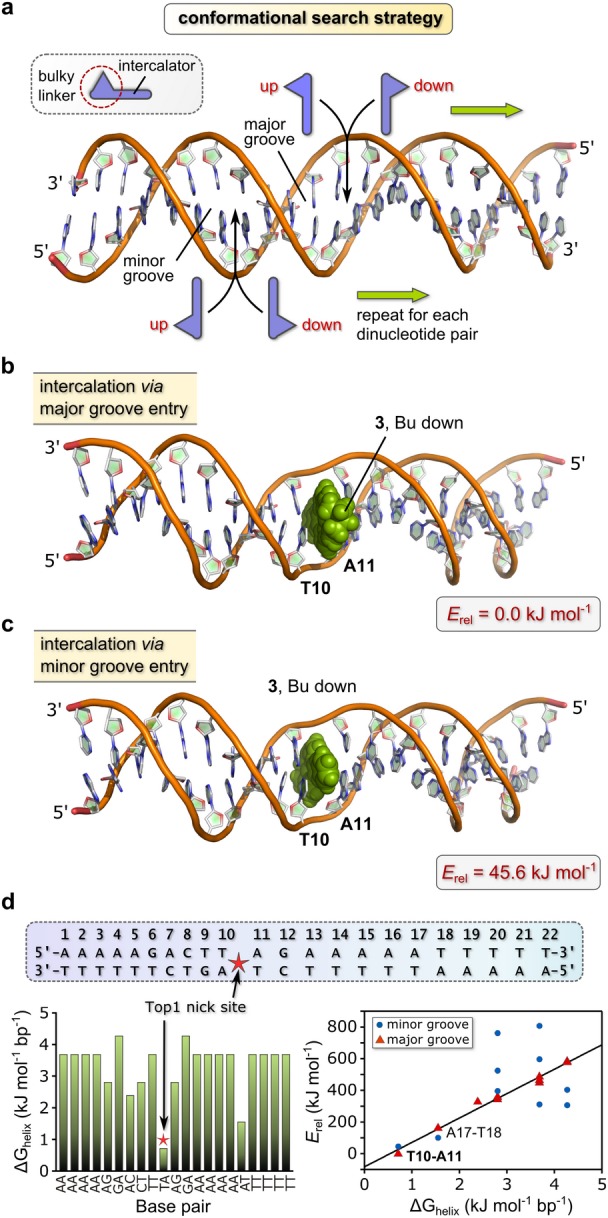
Binding site determination for 3. (a) Conformational search strategy with a 22-bp DNA duplex sequence [Part (d)] favored by Top1 and a modified SP4 force field for macromolecular simulations. A macrocycle such as 3 may intercalate between an adjacent base pair in one of four energetically distinct ways. The search proceeds in a manner akin to stepping down the rungs of a ladder. (b) Structure of the lowest-energy conformation of DNA·3. The compound intercalates at the 5′-TA-3′ site (T10-A11) via major groove entry. (c) Structure of the next-lowest energy conformation (minor groove T10-A11 intercalation adduct) of DNA·3. The higher energy of this adduct reflects, in part, the non-planar conformation for the DNA-bound Au3+ macrocycle relative to the global minimum. (d) Graph of thermodynamic stability vs base pair (left) for the 22-bp DNA duplex. ΔGhelix is the empirical free energy penalty for strand separation at the specified base pair. Right: graph of the relative energies of all simulated DNA·3 intercalation adducts vs ΔGhelix. The best-fit straight line for the major groove adducts is given by: Erel = 154(6) (ΔGhelix) – 83(20) kJ mol–1; R2 = 0.985.
