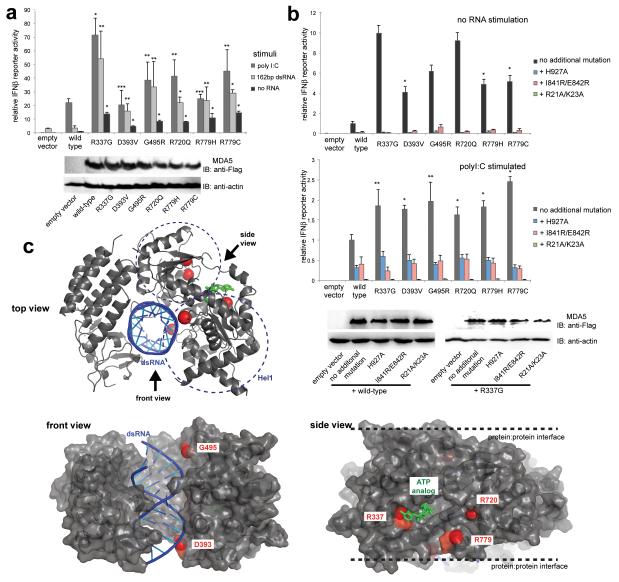
Fig. 3. IFIH1 mutants activate the interferon signaling pathway more efficiently than wild-type IFIH1.
(a) Interferon beta (IFNβ) reporter activity (mean ± SD, n = 3) of Flag-tagged wild-type and mutant IFIH1 with and without stimulation with poly I:C or 162 bp dsRNA in HEK293T cells. The results are representative of three independent experiments. *P<0.005, **P<0.05 and ***P <0.5 (one tailed, unpaired t test, compared with wild-type values). Below are the anti-Flag (F7425, Sigma-Aldrich) and anti-actin (A5441, Sigma-Aldrich) western blots indicating the expression levels of IFIH1 and the internal control (actin), respectively. (b) IFNβ reporter activity (mean ± SD, n = 3) of mutant IFIH1 with and without additional mutations (H927A, I841R/E842R or R21A/K23A) that disrupt RNA binding, filament formation or 2CARD signal activation by IFIH1. Reporter activity was measured in the absence (top) and presence (bottom) of poly I:C stimulation in HEK293T cells. 10 and 20 ng IFIH1 expression constructs were used with and without poly I:C, respectively. The results are representative of three independent experiments. *P<0.005, **P<0.05 (one tailed, unpaired t test). Below are western blots showing the expression levels of wild-type and R337G IFIH1 with and without H927A, I841R/E842R and R21A/K23A. (c) Mapping of the mutated residues (red spheres) onto the structure of IFIH1Δ2CARD (grey) bound by dsRNA (blue) and ATP analog (green). PDB:4GL2.