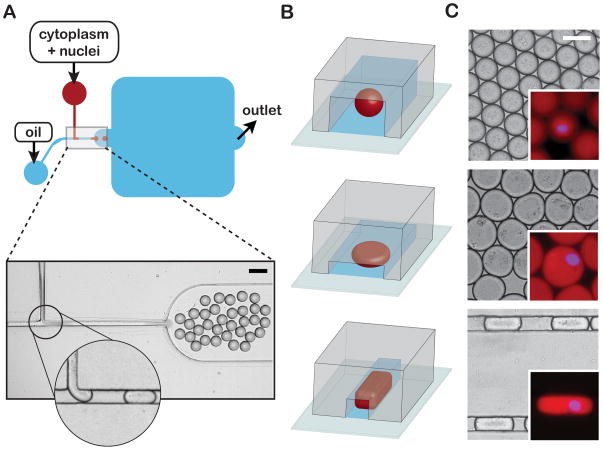
Fig. 1.
Microfluidic encapsulation of nuclei in cytoplasmic volumes of defined size and shape. (A) Top panel - Simplified schematic of a PDMS microfluidic device featuring a classic T-junction droplet generator and collection reservoir (blue rounded-square). An extract phase containing nuclei (red) is continuous until sheared off into droplets by the oil/surfactant phase (blue). Bottom panel - a representative image of droplet generation at a standard T-junction. The magnified inset shows a formed droplet moving toward the collection reservoir and another about to be sheared from the extract phase (see Movie S1). Scale bar = 100 μm. (B) Alternative droplet shapes result from changing the collection reservoir geometry: spheres (top panel), flattened disks (middle panel) and elongated slug-shaped droplets (bottom panel). (C) Representative micrographs of spheres, discs, and slugs contained within reservoirs/channels. Scale bar = 50 μm. Insets: Magnified images of droplets encapsulating “cycled” interphase sperm nuclei. Nuclear DNA was stained with DAPI to access nuclear morphology (blue) and soluble tubulin (pseudo-colored red, labeled with Atto-488) was used to visualize droplet boundaries. All droplets are approximately isovolumetric. Inset scale bar = 50 μm.